Back to Journals » International Journal of General Medicine » Volume 14
The Diagnostic Value of Serum Exosomal Has_circ_0000615 for Breast Cancer Patients
Authors Liu J, Peng X, Liu Y, Hao R, Zhao R, Zhang L, Zhao F, Liu Q, Liu Y, Qi Y
Received 11 May 2021
Accepted for publication 16 July 2021
Published 16 August 2021 Volume 2021:14 Pages 4545—4554
DOI https://doi.org/10.2147/IJGM.S319801
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Jiani Liu,1,2 Xinyu Peng,3 Yinfeng Liu,2 Ran Hao,4 Rumeng Zhao,4 Lei Zhang,4 Fuqing Zhao,4 Qi Liu,4 Yunjiang Liu,1 Yixin Qi1
1Department of Breast Surgery, The Fourth Hospital of Hebei Medical University, Shijiazhuang, 050000, Hebei, People’s Republic of China; 2Department of Breast Surgery, The First Hospital of Qinhuangdao, Qinhuangdao, Hebei, 066000, People’s Republic of China; 3Department of Gastrointestinal Surgery, Affiliated Hospital of Hebei University, Baoding, 071000, Hebei, People’s Republic of China; 4School of Nursing, Hebei Medical University, Shijiazhuang, 050000, Hebei, People’s Republic of China
Correspondence: Yunjiang Liu; Yixin Qi Tel +86 13703297890
; +86 18903113177
Email [email protected]; [email protected]
Background: To explore the expression level of has_circ_0000615 in peripheral blood samples and evaluate its diagnostic value for breast cancer patients.
Methods: The peripheral blood samples of 95 breast cancer patients who underwent curative surgical resection and 95 age-matched healthy volunteers in our institutions from September 2019 to November 2020 were systematically collected. The expression level of has_circ_0000615 in the plasma was amplified and detected by qRT-PCR, and its correlation to clinicopathological characteristics of breast cancer patients were analyzed.
Results: Breast cancer patients had a significantly higher expression level of has_circ_0000615 in the plasma than healthy controls (P < 0.01), and its high expression was closely associated with advanced tumor stage (P=0.010), lymph node metastasis (P = 0.001) and high grade of recurrence risk (P=0.012). The receiver operator characteristic (ROC) curves showed that the area under curve (AUC) value, sensitivity and specificity of has_circ_0000615 for the diagnosis of non-metastatic breast cancer was 0.904 (95% CI: 0.863– 0.944), 76.8% and 88.4%, respectively. Serum has_circ_0000615 expression had a better diagnostic efficiency than routine tumor biomarkers such as CA153, CA125 and CEA for distinguishing breast cancer patients from healthy individuals. TEM revealed that isolated exosomes from the culture medium of breast cancer cells had a disk-like appearance with a diameter of 80– 200 nm vesicles, and the expression of exosome markers CD9 and CD81 was markedly increased. More importantly, the expression of has_circ_0000615 was detected in the exosomes and its expression level was markedly upregulated in breast cancer cell lines compared with normal ductal epithelial cells. The stability assay showed that there was no difference between RNA extraction at 0 hour and 24 hours in terms of the expression of has_circ_0000615 (P =0.327). Has_circ_0000615 might as exosomes be secreted into the circulating blood of breast cancer patients, resulting in a high expression level in plasma samples.
Conclusion: The detection of has_circ_0000615 might be a promising diagnostic method for breast cancer.
Keywords: breast cancer, circRNAs, has_circ_0000615, exosome, diagnostic, biomarker
Introduction
According to the recent Cancer Statistics, breast cancer has become the most prevalent malignancies and a major health problem worldwide in female.1–3 With the advancement in multidisciplinary therapeutic strategy such as surgery, chemoradiotherapy, targeted therapy and endocrine therapy, the prognosis of breast cancer patients has been markedly improved. However, there were still a large proportion of patients dying of breast cancer.4,5 For these patients, the key to successful treatment largely depends on the early diagnosis of tumor. The current data revealed that 5-year survival rate of early-stage breast cancer was more than 90%,5,6 suggesting that the survival outcome of breast cancer patients might be further improved if they were diagnosed at an early stage. Although several serum tumor biomarkers such as carbohydrate antigen 153 (CA153), carcinoembryonic antigen (CEA), carbohydrate antigen 125 (CA125) and carbohydrate antigen 19-9 (CA19-9) were widely used in clinical diagnosis of breast cancer, the sensitivity and specificity of these biomarkers is still limited.7 Therefore, it is urgent to identify novel and effective biomarkers for early diagnosis and prognostic assessment of breast cancer.
It is well established that various components are released into the circulating blood due to cell apoptosis, tumor necrosis or active secretion during the development and cancer progression.8 As a kind of extracellular vesicles secreted by the cells, exosomes played an important role in intercellular communication and various physiological and pathological process such as cellular metabolism.9 With the development of highly sensitive technologies, liquid biopsy made serial biopsies possible for cancer patients through noninvasive approaches.10
Circular RNAs (circRNAs) are a novel class of endogenous non-coding RNAs formed by back-splicing events and characterized by closed continuous loop without polarity and polyadenylate tail structures.11,12 Unlike traditional linear RNAs such as long noncoding RNA (lncRNA) and microRNAs (miRNAs), circRNAs were not degraded by RNases or RNA exonucleases, and were more stable and conserved in peripheral blood or plasma.13,14 It has been reported that circRNAs function as miRNA sponges, RNA-binding proteins or a template of translation into polypeptides, regulating the pathogenesis and progression of various diseases.15–17 Some studies have demonstrated that the expression of circRNAs is markedly dysregulated in tumor samples and widely involved in multiple types of human cancers.18–20 These findings suggested that circRNAs might be an emerging and promising biomarker for the diagnosis and prognostic assessment of malignancies. Recently, a comprehensive microarray and RNA-Seq analysis identified a novel circRNA, has_circ_0000615, which was spliced from the ZNF609 gene, with the length of 874 nt.21 The studies have shown that the expression of has_circ_0000615 was markedly dysregulated in gastric cancer and nasopharyngeal cancer tissues, and knockdown of circ_0000615 inhibited various malignant phenotype of cancer cells.21,22 These findings suggested that has_circ_0000615 might act as a crucial regulator for cancer development and progression. However, its expression level and biological roles in breast cancer remain unknown. By detecting the expression level of has_circ_0000615 in the plasma samples, our previous study demonstrated that has_circ_0000615 was differentially expressed in breast cancer patients compared to healthy controls. Therefore, the current study was further designed to explore the diagnostic value of has_circ_0000615 as serum exosome for breast cancer patients.
Materials and Methods
Study Population and Tissue Samples
In this study, a total of 95 patients who underwent curative surgical resection for primary breast cancer at the fourth hospital of Hebei Medical University between September 2019 and November 2020 were included. All patients did not receive any chemotherapy, radiotherapy, targeted therapy or endocrine therapy before surgery. Those patients with primary underlying diseases such as hypertension, diabetes mellitus, coronary heart disease and the history of other malignancies were excluded. For the purpose of this study, the fasting blood samples of all breast cancer patients were systematically collected before surgery. In addition, 95 age-matched healthy volunteers were recruited as the control populations and blood samples were collected during the same research period. This study was approved by the Ethics Committee of the fourth hospital of Hebei Medical University, and written informed consents were obtained from each study subjects. All procedures were performed in accordance with the Declaration of Helsinki.
RNA Isolation, Reverse Transcription and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Plasma samples of all study subjects were isolated from 4 mL peripheral blood using BD Vacutainer tubes with Ethylene diamine tetraacetic acid (EDTA) as anticoagulation (BD, New Jersey, USA). Blood samples were centrifuged at 1500 rpm for 15 min and the supernatant was transferred to 1.5 mL RNase-free tubes (AXYGEN, JIANGSU, China) and was stored at −80°C until use.
Total RNA in the plasma of breast cancer patients and healthy volunteers were extracted by TIANamp Virus RNA Kit (TIANGE, Beijing) according to the instructions of manufacturer. The purity and concentration of total RNA samples were detected by NanoDrop 1000 instrument spectrophotometer (Thermo Scientific, Waltham, MA, USA). Total RNAs were reversely transcribed into cDNA, and then qRT-PCR assay was performed using SYBR Premix Ex Taq II (Takara, Japan) and the Applied Biosystems Real-Time PCR System. The expression level of has_circ_0000615 in the plasma was determined by qRT-PCR assay using the following sequences of the primers: has_circ_0000615, Forward, 5ʹ- CAGCGCTCAATCCTTTGGGA −3ʹ; Reverse, 5ʹ- GACCTGCCACATTGGTCAGTA −3ʹ. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control (Forward, 5ʹ-TCGACAGTCAGCCGCATCTTCTTT-3ʹ, Reverse, 5ʹ-ACCAAATCCGTTGACTCCGACCTT-3ʹ). The results of qRT-PCR analysis were presented by 2−ΔΔ CT method and the relative expression level of has_circ_0000615 was normalized to the GAPDH expression.
Electrophoresis and Sequencing of qRT-PCR Products
To verify the qRT-PCR products of has_circ_0000615 in the plasma, 5 μL of the product mixture was subjected to 2% agarose gel electrophoresis. The qRT-PCR product of has_circ_0000615 was sent for DNA sequencing at Sangon Biotech Co., Ltd. (Sangon, Shanghai, China).
Detection of Serum CEA, CA125 and CA153
The concentrations of serum tumor biomarkers in breast cancer patients and healthy controls, including CEA, CA125 and CA153, were measured by the Cobas e602 system with the Elecsys CEA Assay kit (Roche Diagnostics, Basel, Switzerland) according to the manufacturer’s instructions. The cutoff values of CEA, CA153 and CA125 were 5 ng/mL, 37 U/mL and 24U/mL, respectively.
Cell Culture and Exosomes Extraction
Human breast cancer cell lines were obtained from the National Institute of Cell Biology (Shanghai, China), and were cultured with Dulbecco’s Modified Eagle Medium (DMEM, Gibco, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin (Gibco) at a 37°C humidified atmosphere containing 5% CO2. Exosomes were extracted from the medium samples using the Transexo Cell Media Exosome Kit (TransGen, Beijing), and all procedures were performed according to the manufacturer’s protocol. We used 1mL of cell supernatant to precipitate the finally obtained exosomes for transmission electron microscopy in the downstream. 6mL of cell supernatant, and the resulting exosomes were used for downstream Western blot and qPCR experiments. For the serum samples, exosomes were isolated from the supernatant using the Plasma Exosome Total RNA Extraction Kit (Transgen, Beijing).
Transmission Electron Microscopy Analysis
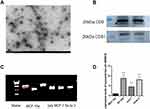
Extracted exosomes were suspended using 30 μL phosphate-buffered saline (PBS) and then were maintained at 4°C. Excess liquid from the edge of the bead was absorbed using filter paper, and then was stained with 1% uranyl acetate dye solution for 1 min. After drying at room temperature, exosomes were visualized by transmission electron microscope (Hitachi ht7800)
Western Blot
Total protein were collected and lysed with radio-immunoprecipitation assay (RIPA), and then they were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Subsequently, the membranes were incubated with monoclonal rabbit anti-CD9 (1:1000, ET1601-9, Huaan Biology) and monoclonal rabbit anti-CD81 antibody (1:1000, ET1611-87, Huaan Biology) at 4°C overnight. Enhanced chemiluminescence (Cell Signaling Technology) was used to visualize the protein bands.
Statistical Analysis
All data and statistical analysis were analyzed using SPSS 22.0 version software package (SPSS, Chicago, IL, USA). In this study, all quantitative data were expressed as mean ± standard deviation (SD) and difference comparisons between the groups were performed by Student’s t-test if the data were accorded with the normal distribution. The relationship between the expression of has_circ_0000615 and clinicopathological features of breast cancer patients were evaluated using chi-square test or Fisher’s exact test. Receiver operator characteristic curve (ROC) was plotted to evaluate the diagnostic value of has_circ_0000615 expression for breast cancer patients. A p-value <0.05 was considered statistically significant difference.
Results
Validation of the Presence of Has_circ_0000615 in the Plasma
To validate the presence of has_circ_0000615 isolated from the blood samples of breast cancer patients, the circRNAs were amplified by qRT-PCR using divergent primers. Agarose electrophoresis assay showed a PCR product with 131kb size of the circRNA fragment (Figure 1A). The Sanger sequencing of the amplified product further revealed that the sequence of has_circ_0000615 had a structure of back-splice junction, which was in accordance with circBase (Figure 1B). These findings suggested that has_circ_0000615 is present in peripheral blood of breast cancer patients and could be detected specifically by qRT-PCR.
The Expression Level of Has_circ_0000615 in the Plasma Samples and Its Correlation to Clinicopathological Characteristics of Breast Cancer Patients
To further determine the expression level of has_circ_0000615, the blood samples of 95 breast cancer patients and 95 age-matched healthy controls were tested. As shown in Figure 2A, breast cancer patients had a significantly higher expression level of has_circ_0000615 in the plasma than healthy controls (P < 0.001). Based on these findings, we investigated the correlation between the expression of has_circ_0000615 and clinicopathological characteristics of 95 breast cancer patients to explore its clinical significance. The results demonstrated that high expression level of has_circ_0000615 was closely associated with advanced tumor stage (P = 0.010), lymph node metastasis (P = 0.001) and high grade of recurrence risk (P = 0.012) (Table 1). On the other hand, we also detected the expression level of has_circ_0000615 in 38 paired breast cancer samples. Consistent with the expression level in the plasma, the expression of has_circ_0000615 was significantly increased in breast cancer tissues compared with adjacent normal tissues (P < 0.001) (Figure 2B).
 |
Table 1 Relationship Between Expression Level of Has_circ_0000615 and Clinical Features in Breast Cancer Patients |
The Diagnostic Value of Has_circ_0000615 in the Plasma for Breast Cancer Patients
Next, we analyzed the ROC curves to evaluate the diagnostic value of serum has_circ_0000615 for breast cancer patients. Our results indicated that the area under curve (AUC) value of has_circ_0000615 was 0.904 (95% CI: 0.863–0.944) for the diagnosis of non-metastatic breast cancer, with the sensitivity of 76.8% and specificity of 88.4%, respectively (Figure 2C). The expression of serum has_circ_0000615 had a better diagnostic efficiency than routine tumor biomarkers for discriminating non-metastatic breast cancer patients from healthy individuals (AUC value for CA153: 0.482, 95% CI: 0.399–0.564, sensitivity: 14.7% and specificity: 92.6%; AUC value for CA125: 0.513, 95% CI: 0.430–0.595, sensitivity: 41.1% and specificity: 67.4%; AUC value for CEA: 0.498, 95% CI 0.415–0.580, sensitivity: 15.8% and specificity: 91.6%). Moreover, we also found that detection of serum has_circ_0000615 had an excellent diagnostic performance for distinguishing early-stage breast cancer (stage 0 and stage I breast cancer diagnosed by histopathology) patients from healthy individuals. The AUC value, sensitivity and specificity of has_circ_0000615 for the diagnosis of early-stage breast cancer were 0.799, 79.2% and 68.7%, respectively, which were better than that of CA153 (AUC:0.383, 95% CI: 0.269–0.497, sensitivity: 14.6% and specificity: 91.7%), CA125 (AUC:0.472, 95% CI 0.356–0.589, sensitivity: 14.6% and specificity: 91.7%) and CEA (AUC:0.475, 95% CI 0.358–0.592, sensitivity: 81.3% and specificity: 25.0%) (Figure 2D).
Exosomal Has_circ_0000615 Might Be Secreted by Breast Cancer Cells into the Plasma
Considering the diagnostic value of has_circ_0000615 in the plasma of breast cancer patients, we further determined whether it was encapsulated in exosomes and secreted into the plasma. For this purpose, we collected the exosomes in the culture medium of breast cancer cells, and transmission electron microscopy revealed that isolated exosomes secreted by breast cancer cells had a disk-like appearance with a diameter of 80–200 nm vesicles (Figure 3A). On the other hand, Western blot analysis showed the presence of the exosome markers CD9 and CD81 in the breast cancer cells (Figure 3B). The expression levels of exosomal has_circ_0000615 was detected in both normal breast ductal epithelial cells and breast cancer cell lines (Figure 3C). Compared with normal ductal epithelial cells, the expression levels of exosomal has_circ_0000615 in MCF-7 cells, Sk-BR-3 cells and 549 cells were markedly upregulated (Figure 3D). What is more, we randomly selected and collected the blood samples of 14 breast cancer patients to compare the expression level of has_circ_0000615 in plasma exosomes and plasma, and the results showed that there was no expression difference between the two groups (P = 0.243) (Figure 4A). The stability assay showed that there was no difference between RNA extraction at 0 hour and 24 hours in terms of the expression of has_circ_0000615 (P = 0.327) (Figure 4B). These findings suggested that has_circ_0000615 might as exosomes be secreted by breast cancer cells into the plasma.
Discussion
Recently, increasing evidence have showed that aberrant expression of circRNAs is implicated in the carcinogenesis and tumor progression. For instance, a previous study reported that the expression of CircZNF609 was significantly increased in breast cancer tissues and cell lines, and its high expression was a predictor of poor prognosis.23 Huang et al reported a low expression level of has-circ-0000745 in gastric cancer tissues in comparison to adjacent normal tissues, and its expression level was proved to be a promising diagnostic biomarker for this malignancy.24 In addition to the aberrant expression in tumor tissue samples, circRNAs were encapsulated in exosomes and secreted by cancer cells into the plasma. Yin et al demonstrated that the expression level of has-circ-0001785 in the plasma was markedly higher in breast cancer patients than in healthy controls, and high has-circ-0001785 expression in the plasma was closely associated with histological grade, TNM stage and distant metastasis.25 More importantly, has-circ-0001785 showed a better diagnostic value for breast cancer than other tumor biomarkers such as CA153 and CEA.25
Unlike lncRNA, miRNA or other linear RNAs, the biological stability of circRNAs was more suitable in the plasma, making them as desirable biomarkers for the diagnosis of human diseases such as cancers. In the present study, we identified and detected the expression level of has_circ_0000615 in the plasma using qRT-PCR analysis. The results revealed that the expression level of has_circ_0000615 was markedly upregulated in breast cancer patients compared with age-matched healthy individuals. Based on the correlation between has_circ_0000615 expression and clinicopathologic features, we found that the expression of has_circ_0000615 was higher in breast cancer patients with advanced tumor stage, lymph node metastasis and high recurrence risk. Considering the dysregulated expression of has_circ_0000615 in the plasma and its clinical significance, we also evaluated the diagnostic efficiency of has_circ_0000615 for breast cancer patients. Unsurprisingly, the detection of has_circ_0000615 in the plasma properly distinguished breast cancer patients from healthy individuals. ROC curves suggested that plasma has_circ_0000615 could be used as a more desirable biomarker for the diagnosis of breast cancer patients, compared with CA153, CA125 and CEA.
It has been well established that the circRNAs could be packaged by exosomes and be released into the circulating blood.26 To date, the circRNAs in exosomes have been considered as potential diagnostic and prognostic biomarkers for human cancers and been paid more attention.27–29 In the current study, we collected the isolated exosomes from breast cancer cells and used transmission electron microscopy to validate the appearance and diameter of exosomes. Moreover, the expression of CD9 and CD81 protein was markedly elevated in breast cancer cell lines, further revealing the presence of the exosomes in culture medium. Interestingly, we also found that breast cancer cell lines had a higher expression level of has_circ_0000615 in the exosomes than normal breast ductal epithelial cells. These findings revealed that has_circ_0000615 might be encapsulated in exosomes and secreted by breast cancer cells.
Although the dysregulation of has_circ_0000615 expression and its diagnostic value for breast cancer patients has been validated by our study, its biological function in the tumorigenesis and progression of breast cancer remain unclear. In a recent study, Liu et al reported that has_circ_0000615 was markedly overexpressed in nasopharyngeal cancer tissues and cell lines, and the malignant phenotypes of nasopharyngeal cancer cells were significantly suppressed by has_circ_0000615 knockdown.22 Their study further revealed that the biological behaviors of nasopharyngeal cancer cells were inhibited by has_circ_0000615 knockdown via regulating the miR-338-3p/FGF2 axis.22 In the future, further investigations need to be performed to explore the effects of has_circ_0000615 on the cell growth, migration and invasion ability of breast cancer and underlying molecular mechanisms.
In conclusion, high expression level of has_circ_0000615 was markedly detected in both peripheral blood samples and cell lines of breast cancer, and was significantly correlated with aggressive clinicopathological features of tumor. Has_circ_0000615 might be packaged by exosomes and be secreted into the circulating blood. These findings suggested that has_circ_0000615 was a promising biomarker for the diagnosis of breast cancer.
Data Sharing Statement
All data generated or analyzed during this study are available from the corresponding author Yunjiang Liu upon reasonable request.
Ethical Approval
This study was approved by the Ethics Committee of the Fourth Hospital of Hebei Medical University, and written informed consents were obtained from each study subjects.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was financially supported by the Foundation of Key R&D Program of Hebei Province (No. 19277799D) and Science and Technology R&D Program of Qinhuangdao (No. 201805A158).
Disclosure
No financial or nonfinancial benefits have been received or will be received from any party related directly or indirectly to the subject of this article.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi:10.3322/caac.21654
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi:10.3322/caac.21590
3. Li N, Deng Y, Zhou L, et al. Global burden of breast cancer and attributable risk factors in 195 countries and territories, from 1990 to 2017: results from the Global Burden of Disease Study 2017. J Hematol Oncol. 2019;12:140. doi:10.1186/s13045-019-0828-0
4. DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69:438–451. doi:10.3322/caac.21583
5. Kang SY, Kim YS, Kim Z, et al. Breast cancer statistics in Korea in 2017: data from a breast cancer registry. J Breast Cancer. 2020;23:115–128. doi:10.4048/jbc.2020.23.e24
6. Zhang J, Zhao B, Jin F. The assessment of 8th edition AJCC prognostic staging system and a simplified staging system for breast cancer: the analytic results from the SEER database. Breast J. 2019;25:838–847. doi:10.1111/tbj.13347
7. Wang W, Xu X, Tian B, et al. The diagnostic value of serum tumor markers CEA, CA19-9, CA125, CA15-3, and TPS in metastatic breast cancer. Clin Chim Acta. 2017;470:51–55. doi:10.1016/j.cca.2017.04.023
8. Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665.
9. Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi:10.1016/j.imlet.2006.09.005
10. Riethdorf S, O’Flaherty L, Hille C, Pantel K. Clinical applications of the cell search platform in cancer patients. Adv Drug Deliv Rev. 2018;125:102–121.
11. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi:10.1038/nbt.2890
12. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi:10.1038/nature11928
13. Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15:9331–9342. doi:10.3390/ijms15069331
14. Wilusz JE. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA. 2018;9:e1478. doi:10.1002/wrna.1478
15. Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi:10.1016/j.molcel.2014.08.019
16. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi:10.1038/nature11993
17. Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi:10.1016/j.molcel.2013.08.017
18. Wu G, Zhou W, Pan X, et al. Circular RNA profiling reveals exosomal circ_0006156 as a novel biomarker in papillary thyroid cancer. Mol Ther Nucleic Acids. 2020;19:1134–1144. doi:10.1016/j.omtn.2019.12.025
19. Chen W, Cen S, Zhou X, et al. Circular RNA CircNOLC1, upregulated by NF-KappaB, promotes the progression of prostate cancer via miR-647/PAQR4 axis. Front Cell Dev Biol. 2020;8:624764. doi:10.3389/fcell.2020.624764
20. Cen J, Liang Y, Huang Y, et al. Circular RNA circSDHC serves as a sponge for miR-127-3p to promote the proliferation and metastasis of renal cell carcinoma via the CDKN3/E2F1 axis. Mol Cancer. 2021;20:19. doi:10.1186/s12943-021-01314-w
21. Dong Z, Liu Z, Liang M, et al. Identification of circRNA-miRNA-mRNA networks contributes to explore underlying pathogenesis and therapy strategy of gastric cancer. J Transl Med. 2021;19:226. doi:10.1186/s12967-021-02903-5
22. Liu HS, Zheng RN, Guo LB, Fu XJ. Circular RNA circ_0000615 knockdown suppresses the development of nasopharyngeal cancer through regulating the miR-338-3p/FGF2 axis. Neoplasma. 2020;67:1032–1041. doi:10.4149/neo_2020_191019N1061
23. Wang S, Xue X, Wang R, et al. CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. Cancer Manag Res. 2018;10:3881–3890. doi:10.2147/CMAR.S174778
24. Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23:6330–6338. doi:10.3748/wjg.v23.i34.6330
25. Yin WB, Yan MG, Fang X, Guo JJ, Xiong W, Zhang RP. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta. 2018;487:363–368. doi:10.1016/j.cca.2017.10.011
26. Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi:10.1016/j.omtn.2019.02.005
27. Wang S, Zhang K, Tan S, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer. 2021;20:13. doi:10.1186/s12943-020-01298-z
28. Lyu D, Huang S. The emerging role and clinical implication of human exonic circular RNA. RNA Biol. 2017;14:1000–1006. doi:10.1080/15476286.2016.1227904
29. Lyu L, Zhang S, Deng Y, et al. Regulatory mechanisms, functions, and clinical significance of CircRNAs in triple-negative breast cancer. J Hematol Oncol. 2021;14:41. doi:10.1186/s13045-021-01052-y
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.