Back to Journals » Cancer Management and Research » Volume 12
The Clinical Relevance and Function of Krüppel-Like Factor 16 in Breast Cancer
Authors Bang S , Li J, Zhang M , Cui R, Wu X, Xin Z, Ma D, Zhang J, Zhang H
Received 1 April 2020
Accepted for publication 25 June 2020
Published 27 July 2020 Volume 2020:12 Pages 6373—6383
DOI https://doi.org/10.2147/CMAR.S256490
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Bilikere Dwarakanath
Soyeon Bang,1 Junhong Li,2 Meiqin Zhang,3 Renjie Cui,3 Xingwen Wu,4 Zhaochen Xin,1 Duan Ma,3,5 Jin Zhang,3 Hongwei Zhang1
1Department of General Surgery, Zhongshan Hospital, Fudan University, Shanghai, People’s Republic of China; 2Department of Urology, Shanghai Fifth People’s Hospital, Fudan University, Shanghai, People’s Republic of China; 3Key Laboratory of Metabolism and Molecular Medicine, Ministry of Education, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Institutes of Biomedical Sciences, Shanghai Medical College, Fudan University, Shanghai, People’s Republic of China; 4Department of Dentistry, Zhongshan Hospital, Fudan University, Shanghai, People’s Republic of China; 5Children’s Hospital, Fudan University, Shanghai, People’s Republic of China
Correspondence: Hongwei Zhang
Department of General Surgery, Zhongshan Hospital, Fudan University, Shanghai 200032, People’s Republic of China
Tel +86-13681973106
Fax +86 21-64038472
Email [email protected]
Duan Ma
Key Laboratory of Metabolism and Molecular Medicine, Ministry of Education, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Institutes of Biomedical Sciences, Shanghai Medical College, Fudan University, 130 Dong an Road, Xuhui District, Shanghai 200032, People’s Republic of China
Tel +86-13916126452
Fax +86 21-54237441
Email [email protected]
Background: Krüppel-like factor 16 (KLF16), a member of the KLF family, is involved in metabolism and regulation of the endocrine system and has emerging roles in tumor progression. However, the expression of KLF16 and its role in breast cancer are elusive.
Methods: We investigated the expression and prognostic value of KLFs in breast cancer using data acquired from the TCGA BRCA dataset and the Kaplan–Meier plotter dataset. The protein levels of KLF16 in breast specimens were detected by immunohistochemistry (IHC). KLF16 silencing using shRNAs was performed to explore the effects of KLF16 on breast cancer cell growth, migration, and invasion. The expression of EMT markers in cells manipulated for KLF16 expression was assessed by Western blotting.
Results: Using publicly available dataset and specimens from breast cancer patients, we found that the expression levels of KLF16 were significantly higher in tumor tissues and that high levels of KLF16 were associated with poor prognosis in breast cancer patients. Moreover, KLF16 expression levels had relation to several clinicopathological parameters of breast cancer, including the molecular subtype and histological grade. Importantly, knockdown of KLF16 dramatically suppressed cell proliferation both in vitro and in vivo. Also, KLF16 deletion impaired migration, and invasion in breast cancer cells, and suppressed epithelial–mesenchymal transition (EMT).
Conclusion: Our results suggest that KLF16 has important oncogenic functions in breast cancer and that the expression levels of KLF16 are associated with prognosis in breast cancer patients. Our findings also suggest that KLF16 is involved in proliferation, migration, and invasion in breast cancer cells. Thus, KLF16 might be a promising prognostic marker and a therapeutic target for breast cancer.
Keywords: breast cancer, EMT, Krüppel-like factor 16, metastasis, proliferation
Introduction
Breast cancer is one of the most common female cancers in the world. With 2,088,849 estimated new cases and 626,679 breast cancer-related deaths in 2018, breast cancer is the most commonly diagnosed cancer and the leading cause of cancer-related death among global women.1 Similarly, the incidence of breast cancer ranks first in women in China and South Korea.2,3 Breast cancer is a heterogeneous disease that can be classified into several subtypes based on histological characteristics and the expression of genetic markers.4 Despite recent advances in breast cancer therapy, including chemotherapy, endocrine therapy, and HER2-targeted therapies,5–7 the high proliferative, invasive, metastatic potential of breast cancer cells, as well as the development of drug resistance, are major causes of therapeutic failure and poor survival in patients with breast cancer.8,9 Therefore, the development of novel therapeutic approaches is an urgent need.
The Krüppel-like factor (KLF) family includes at least 17 members, all of which are zinc finger-containing transcription factors. KLFs bind to GC-rich DNA regions and regulate gene expression, thereby playing vital roles in multiple biological processes, such as cell proliferation, apoptosis, migration, and differentiation.10 Numerous studies have reported that KLFs serve as tumor suppressors or oncogenes depending on the specific cellular context.11 Mounting evidence also suggests that KLF members are involved in the development and progression of breast cancer. For example, the long non-coding RNA (lncRNA) RUSC1-AS1 has been shown to promote breast cancer progression by epigenetic silencing of KLF2.12 KLF3 downregulation has been shown to inhibit the migration and invasion of breast cancer cells by promoting STAT3 protein expression.13 KLF4 has been implicated in breast cancer development by regulating estrogen signaling,14 while KLF5 has different functions in different types of breast cancer. In basal-like breast cancer, a super-enhancer drives KLF5 upregulation, which subsequently drives cell proliferation, migration, and stemness.15 On the other hand, in estrogen receptor-positive (ER+) breast cancer, KLF5 inhibits cell proliferation by suppressing the transcriptional activity of ERα.16 KLF6 inhibits estrogen receptor-mediated breast cancer cell growth via a c-Src-mediated pathway.17 However, A KLF6 splice variant, KLF6-SV1 promotes an EMT-like phenotype and increases cancer cell invasion in part via TWIST1.18 KLF8 has also been shown to promote metastasis in breast cancer by regulating matrix metallopeptidase 9 (MMP9),19 as well as by activating MMP14 in cooperation with FAK.20 KLF9 and KLF17 have also been implicated in epithelial-mesenchymal transition (EMT) and metastasis in breast cancer.21,22 Klf10 induces breast cancer cell apoptosis through modulation of BI-1 expression and inhibits breast cancer invasion and metastasis by inhibition of EGFR transcription and the EGFR Signaling Pathway.23,24 miR-30d promotes breast cancer cell growth, metastasis and EMT by targeting KLF11.25 KLF12 downregulation by miR-205 inhibits invasion and promotes apoptosis in basal-like breast carcinoma.26 KLF14 transcription is significantly reduced in breast cancer and is significantly negatively correlated with Plk4.27 Tumor-suppressive roles have been attributed to KLF15 in breast cancer cells, which are mediated by the induction of p21 and the subsequent inhibition of cell cycle progression.28 However, no studies have examined the role of KLF16 in human breast cancer.
In the present study, we found that KLF16 expression was upregulated in breast cancer tissues, both in The Cancer Genome Atlas (TCGA) BRCA Dataset and in specimens acquired from breast cancer patients. High levels of KLF16 are associated with poor disease-free survival (DFS) rates in breast cancer patients. KLF16 silencing significantly repressed tumor growth in breast cancer cells, as well as severely impaired their migration and invasion. Our study demonstrated that KLF16 plays oncogenic roles in breast cancer and that it might be a potential therapeutic target.
Materials and Methods
Cell Culture
The human breast cancer cell lines MCF-7 and MDA-MB-231 were obtained from the Stem Cell Bank (Chinese Academy of Sciences, Shanghai, China). Both cell lines were maintained in DMEM (Corning Inc., Corning, NY, USA) supplemented with 10% fetal bovine serum (Cegrogen Biotech, A0500-3010, Stadtallendorf, Germany) and 1% penicillin/streptomycin (BI, 03-031-1BCS, Israel), in a 5% CO2 humidified incubator at 37°C.
Plasmids and Lentiviral Infections
The short hairpin RNA (shRNA) sequences used were as follows:
shKLF16 #1 (5′-CTCGCACCTAAAGTCGCACCT-3′);
shKLF16 #2 (5′-CTACAAGTCCTCGCACCTAAA-3′);
shSCR (5′-GCTCCGTGAACGGCCACGAGT-3′); all sequences were cloned into the pLKO.1 vector.
HEK293FT cells were transfected with shRNA-pLKO.1 plasmid using PEI 25K (Polysciences, 23966-1, Warrington, PA, USA) according to the manufacturer’s instructions. MCF-7 and MDA-MB-231 cells were transduced with the lentivirus, and stable cell lines were generated after selection with puromycin (Amresco, 1328-J593-25MG, Albany, NY, USA) for seven days.
Cell Growth Assay
The proliferation of MCF-7 and MDA-MB-231 cells was assessed using the Cell Counting Kit-8 (Dojindo, CK04, Kumamoto, Japan). A total of 1000 cells/well were seeded into 96-well plates. After 24 hours of incubation, CCK8 reagent was added into 100 µL of DMEM medium containing 10% FBS and cells were incubated at 37°C for one hour. After incubation, the absorbance at 450nm was measured using a microplate reader. This process was repeated for four days.
Colony Formation Assay
Cells (1000 cells/well) were seeded in 6-well plates in DMEM medium supplemented with 10% FBS. Seven days later (when colonies had reached an appropriate size), the cells in each well were fixed with methanol for 10 minutes and stained with 0.1% crystal violet for 25 minutes.
Western Blotting Analysis
Cells were lysed in lysis buffer containing protease inhibitor cocktail (Roche Diagnostics), and the concentrations of the extracted proteins were measured using the Pierce BCA Protein Assay Kit (Thermo Fisher, 23225, Waltham, MA, USA). Protein samples were separated by SDS-PAGE, transferred onto nitrocellulose membranes (BioTrace™, 66485 NT, Port Washington, NY, USA), and subsequently probed with primary antibodies overnight at 4°C. Next, the membranes were incubated with HRP-conjugated secondary antibodies for one hour to detect the immunoreactive bands. The following antibodies were used: KLF16 (Sigma-Aldrich, HPA052481, St. Louis, MO, USA), β-actin (Proteintech, 20536-1-AP, Wuhan, Hubei, China), Zeb1 (CST,3396, Danvers, Massachusetts, USA), E-cadherin (Bioscience, 1610182, Franklin Lakes, New Jersey, USA), MMP9 (Santa Cruz, SC-10737, Dallas, Texas, USA), and Slug (CST, 9585, Danvers, Massachusetts, USA).
Migration and Invasion Assays
Transwell inserts (Corning Inc., Corning, NY, USA) were used for the migration and invasion assays. To determine cell migration, MCF-7 and MDA-MB-231 cells (2.5×104/well) were seeded on the upper chamber in DMEM (without FBS), and 700 µL of DMEM supplemented with 10% FBS was added to the lower chamber. After 24–48h, transwell inserts were washed with PBS, and cells were fixed with methanol and stained with 0.1% crystal violet. The cells on the upper chamber were removed using a cotton swab. For the invasion assay, the surface of the transwell insert was coated with matrigel (Corning Inc., Corning, NY, USA) diluted (1:8) in DMEM. After the matrigel had solidified, cells (3.0×104/well) were seeded in the upper chamber. From this point, the procedure followed was the same as in the migration assay.
Animal Experiments
The animal experimental procedures in this study were approved by the Institutional Animal Care and Use Committee of Fudan University. All the animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and regulations for Laboratory Animal Science Center, Fudan University which include items for the welfare of the animals. Each 6-week-old male BALB/c nude mice (n=5) was injected subcutaneously with a total of 1×106 Lentivirus-infected MDA-MB-231 cells suspended in Matrigel (volume, 1:1; Corning, Inc., 356234, NY, USA). 28 days later, mice were euthanized and sections of tumor xenografts were dissected and weighed.
Patients and Tissues
One hundred eighty-two surgical specimens from patients with invasive ductal carcinoma (117 breast cancer and 65 adjacent normal tissues) were used in this study. The specimens were obtained from the surgical pathology files of the Zhongshan Hospital, Fudan University, Shanghai, China. None of the patients had received neoadjuvant therapy, such as chemotherapy, radiotherapy, or endocrine therapy. All the tissue specimens were fixed with formalin and embedded in paraffin wax. The research protocol was approved by the Ethics Committee of the Zhongshan Hospital, Fudan University. All patients were informed completely and signed informed consent. All experiments were conducted following the guidelines and regulations of Medical Ethics Committee of Zhongshan Hospital, Fudan University and were conducted in accordance with the Declaration of Helsinki.
Immunohistochemistry (IHC)
The tissue microarray (TMA) slides were deparaffinized, rehydrated, and dipped in 3% hydrogen peroxide for 10 minutes at room temperature. Antigen retrieval was performed in 0.01mol/L citric buffer (pH 6.0) at 95°C for 15–20minutes. The TMA slides were cooled down for 20 minutes before being blocked with 10% BSA. Staining with the KLF16 antibody (HPA052481; Sigma-Aldrich, St. Louis, MO, USA) was conducted overnight at 4°C. Each sample was scored for the percentage (0, 0%; 1, 1%‐25%; 2, 26%‐50%; 3, 51%‐75%; 4, 76%‐100%) and the intensity (0, negative; 1, low; 2, medium; 3, high) of positive cells. IHC scores were determined as the percentage score × intensity score.
Published Datasets and Analysis
The TCGA expression profiles of breast cancer patients were downloaded using the UCSC Xena browser (http://xena.ucsc.edu/). The Kaplan-Meier plotter (www.kmplot.com) was used to assess the association between KLF16 mRNA expression and post-progression survival (PPS) in breast cancer patients.29
Statistical Analysis
Student’s t-test and Mann–Whitney test were used for the comparison of quantitative data between two or more groups. All data values are represented as the mean ± standard deviation based on at least three independent experiments. All data analyses were performed using GraphPad Prism 7 and *p <0.05, **p <0.01, ***p <0.001 was considered statistically significant.
Results
KLF16 Expression Levels are Elevated in Breast Cancer and are Associated with Poor Survival
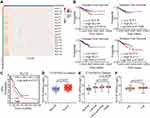
To identify potential oncogenes among KLF family members, we assessed the expression of KLFs between normal and tumor tissues in the TCGA BRCA dataset and found that all KLFs were significantly differentially expressed between normal and tumor tissues (normal, n=114; tumor, n=1086) (Figure 1A). Among them, the roles of KLF1, KLF7, KLF13, and KLF16 in breast cancer have not been previously reported. Survival analysis of these KLFs in the TCGA BRCA dataset revealed that higher KLF16 expression levels were associated with poor disease-free survival (DFS) in patients with breast cancer. Quantile cut-off values were used to analyze disease-free survival (low, n=227; high n=227) (Figure 1B). We also found that high KLF16 expression levels (226328_at) in breast cancer tissues were significantly connected with poor post-progression survival (PPS) (low, n=170; high, n=164) (Figure 1C). Analysis of mRNA levels in breast tissues in the TCGA BRCA dataset revealed that KLF16 mRNA levels were markedly higher in tumor tissues compared to adjacent non-tumor tissues (normal, n=114; tumor, n=1086) (Figure 1D). Furthermore, KLF16 was expressed at higher levels in TNBC breast cancer than in Luminal and HER2-enriched breast cancer (normal, n=114; luminal, n=660; HER2-enriched, n=38; TNBC, n=177) (Figure 1E). HER2-enriched breast cancer did not include all the HER2+ breast cancers and we have excluded the hormone receptor-positive HER2+ patients from the HER2-enriched subtype. Additionally, patients younger than 45 had lower levels of KLF16 compared with patients older than 45 (≤45, n=27; >45, n=908) (Figure 1F).
KLF16 Expression is Associated with Clinicopathological Parameters in Breast Cancer
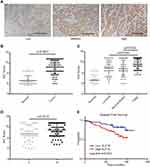
To evaluate further the clinical significance of KLF16 expression in human breast cancer, we performed IHC staining in a human breast tissue microarray (TMA) (Figure 2A). In accordance with the findings from the TCGA BRCA dataset, we found that KLF16 was expressed at higher levels in tumor tissues compared with adjacent normal tissues (normal, n=65; tumor, n=117) (Figure 2B), and the expression of KLF16 in TNBC specimens was significantly higher than in samples from luminal and HER2-enriched subtypes of breast cancer (normal, n=65; luminal, n=33; HER2-enriched, n=29; TNBC, n=55) (Figure 2C). KLF16 is more highly expressed in breast cancer tissues with histological grade III than that in breast cancer tissues with histological grade II (II, n=46; III, n=71) (Figure 2D). Additionally, we also found that high KLF16 expression levels were associated with reduced DFS rate (low, n=48; high, n=69) (Figure 2E). As shown in Table 1, the KLF16 expression levels were related to patient age, histological grade, ER status, PR status, and Ki-67 status. However, no significant association was identified between the KLF16 expression levels and tumor size, lymph node metastasis, or HER-2 status.
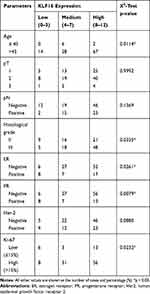 |
Table 1 Association Between KLF16 Expression and Clinicopathological Parameters in Breast Cancer Tissues (n=117) |
KLF16 Promotes Breast Cancer Cell Proliferation in vitro and in vivo
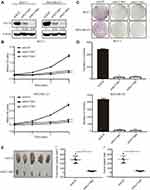
To investigate the role of KLF16 in breast cancer cell proliferation, we silenced KLF16 expression in MCF-7 and MDA-MB-231 cells using two individual shRNAs. KLF16 knockdown was confirmed by Western blotting (Figure 3A). CCK-8 assay indicated that the KLF16 knockdown resulted in a profound decrease in cell proliferation in both cell lines. Compared to the control group (shSCR), the relative cell growth rates of two KLF16 knockdown groups (shKLF16#1 and shKLF16#2) decreased 4.2 and 3.7 fold respectively in MCF-7 cells (Figure 3B). And likewise in MDA-MB-231 cells, the relative cell growth rates of two KLF16 knockdown groups reduced 2.1 and 2.4 fold, respectively (Figure 3B). Furthermore, compared with shSCR-expressing control cells, the colony formation assays showed that KLF16 downregulation by shKLF16#1 and shKLF16#2 significantly reduced cell colony formation in MCF7 cells (26.2 and 23.4 fold, respectively) and in MDA-MB-231 cells (20.6 and 25.9 fold, respectively) (Figure 3C and D). To assess the role of KLF16 in breast cancer cells growth in vivo, the animal models were established. We subcutaneously injected MDA-MB-231 cells infected with shSCR or shKLF16#2 into male athymic nude mice (n=5). Tumors were formed within a month and we measured the xenografts. Compared to the control groups, the size and weight of xenografts were significantly smaller (8.0 fold) and lighter (8.2 fold) in the KLF16-depleted groups (Figure 3E–G). Collectively, these results suggest a crucial role for KLF16 in the cell growth and proliferation of breast cancer cells in vitro and in vivo.
KLF16 Silencing Suppresses Breast Cancer Cell Migration and Invasion
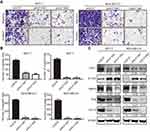
To explore the potential involvement of KLF16 in regulating the metastatic potential of breast cancer cells, we performed transwell migration and invasion assays. The results showed that KLF16 deletion significantly impaired the cell migration and invasion abilities of breast cancer cells (Figure 4A). In MCF-7 cells, KLF16 knockdown with shKLF16#1 and shKLF16#2 significantly diminished cell migration (3.1 and 4.0 fold, respectively) and invasion (15.8 and 17.6 fold, respectively) (Figure 4B). Also, KLF16 disruption with the two shRNAs significantly inhibited migration (13.0 and 30.0 fold, respectively) and invasion (19.4 and 30.2 fold, respectively) in MDA-MB-231 cells (Figure 4B). As EMT is a key process promoting cancer metastasis,30 we explored the effects of KLF16 silencing on the protein levels of EMT markers, including ZEB1, E-cadherin, MMP9, and Slug (Figure 4C). We found that KLF16 deletion resulted in a profound decrease in the protein levels of ZEB1, MMP9, and Slug, while the protein levels of the epithelial marker E-cadherin were increased. These results suggest that KLF16 potentially has a critical pro-metastatic role in breast cancer cells.
Discussion
There have been a number of reports regarding the roles of KLFs in cancer.11 In particular, KLFs have been found to be crucial for the development of breast cancer. In TCGA BRCA dataset, the mRNA levels of all KLFs were significantly differentially expressed between breast cancer tissues and adjacent non-tumor tissues. Previous studies indicated that many KLFs are involved in the development of breast cancer.12–28 In this study, using in silico analyses, we found that KLF16 might have tumor-promoting roles in breast cancer.
KLF16 was first identified as a dopamine receptor-regulating factor involved in the regulation of dopamine transmission in the brain.31 KLF16 has also been shown to suppress adipogenesis by downregulating the expression of PPARγ.32 Moreover, studies have also linked KLF16 to metabolism and endocrine function regulation.33 Emerging studies have revealed that KLF16 plays a major role in regulating tumor cell proliferation and survival.34–37 However, whether KLF16 acts as an oncogene or tumor suppressor is yet controversial. It may depend on the specific cellular context. In human glioma, tumor-suppressive roles have been attributed to KLF16, by regulating TFAM expression.34 However, KLF16 has been demonstrated to promote gastric cancer cell proliferation by regulating cell cycle progression.35 Zhang et al36 reported that KLF16 knockdown suppressed prostate cancer cell growth in vitro and in vivo, and a deficiency of KLF16 inhibited activation of MYC signaling. Besides, KLF16 was highly upregulated in anaplastic thyroid carcinoma.37 In the present study, our findings indicate that KLF16 was associated with proliferation, migration, and invasion abilities of breast cancer cells. Overall, the results of these studies indicated that KLF16 plays diverse roles in tumors.
To our best of knowledge, this is the first study to assess the expression of KLF16 in breast cancer tissues and to investigate its biological significance in breast cancer cells. Our study demonstrated that KLF16 has tumor-promoting roles in breast cancer and is associated with adverse clinical outcomes. We found that KLF16 expression was significantly upregulated in breast cancer tissues compared to adjacent normal tissues, especially in TNBC, which had the highest expression of KLF16. We also found that high KLF16 expression was associated with reduced DFS rate, both in the TCGA BRCA dataset and in clinical specimens from breast cancer patients. The worse DFS rate in high KLF16 expression group may result from enrichment of TNBC subtype which is associated with poor clinical outcomes.38,39 Furthermore, apart from KLF16 deletion inhibited breast cancer cells growth in vitro, targeting KLF16 could depress the growth of breast cancer cells in vivo. Therefore, specific inhibitor for KLF16 in the future may be a potential breast cancer treatment new strategy. And we also found that the deletion of KLF16 obviously suppressed migration, and invasion in MCF-7 and MDA-MB-231 cells. During EMT, epithelial cells acquire a mesenchymal phenotype, which is essential for metastasis.40 Several KLFs, including KLF2, KLF5, and KLF11, have been shown to have a role in EMT in breast cancer.25,41,42 Our findings first demonstrated that KLF16 silencing significantly suppressed EMT, as indicated by the decreased protein levels of ZEB1, MMP9, and Slug and the higher levels of the epithelial marker E-cadherin. However, the detailed mechanisms by which KLF16 promotes EMT and metastasis need to be further investigated.
In conclusion, the findings of this study suggested that KLF16 may play important roles in the development and progression of breast cancer, highlighting it as a potential therapeutic target. Therefore, understanding the mechanisms underlying the effects of KLF16 in the progression of breast cancer, as well as the precise role of KLF16 in EMT, may facilitate the development of novel therapeutic approaches against the progression of breast cancer.
Abbreviations
TCGA, The Cancer Genome Atlas; BRCA, breast cancer susceptibility gene; KLF, Krüppel-like factor; EMT, epithelial–mesenchymal transition; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor Receptor-2; TNBC, triple negative breast cancer; E-cadherin, epithelial cadherin; MMP9, matrix metallopeptidase 9; SLUG, snail family zinc finger2; ZEB-1, zinc finger E-box-binding homeobox 1; IHC, immunohistochemistry.
Acknowledgments
We would like to acknowledge the support and assistance of the members of the Department of General Surgery (Breast) and Department of Pathology, Zhongshan Hospital, Fudan University, Shanghai, China.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
3. Jung KW, Won YJ, Kong HJ, Lee ES. Prediction of cancer incidence and mortality in Korea, 2019. Cancer Res Treat. 2019;51(2):431–437. doi:10.4143/crt.2019.139
4. Di Cosimo S, Baselga J. Management of breast cancer with targeted agents: importance of heterogeneity. [corrected]. Nat Rev Clin Oncol. 2010;7(3):139–147. doi:10.1038/nrclinonc.2009.234
5. Bartlett JMS, Sgroi DC, Treuner K, et al. Breast Cancer Index and prediction of benefit from extended endocrine therapy in breast cancer patients treated in the Adjuvant Tamoxifen-To Offer More? (aTTom) trial. Ann Oncol. 2019;30(11):1776–1783. doi:10.1093/annonc/mdz289
6. Braso-Maristany F, Griguolo G, Pascual T, et al. Phenotypic changes of HER2-positive breast cancer during and after dual HER2 blockade. Nat Commun. 2020;11(1):385. doi:10.1038/s41467-019-14111-3
7. Gray E, Marti J, Wyatt JC, Brewster DH, Hall PS; group Sa. Chemotherapy effectiveness in trial-underrepresented groups with early breast cancer: a retrospective cohort study. PLoS Med. 2019;16(12):e1003006. doi:10.1371/journal.pmed.1003006
8. Bertucci F, Ng CKY, Patsouris A, et al. Genomic characterization of metastatic breast cancers. Nature. 2019;569(7757):560–564. doi:10.1038/s41586-019-1056-z
9. Formisano L, Lu Y, Servetto A, et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat Commun. 2019;10(1):1373. doi:10.1038/s41467-019-09068-2
10. McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90(4):1337–1381. doi:10.1152/physrev.00058.2009
11. Tetreault MP, Yang Y, Katz JP. Kruppel-like factors in cancer. Nat Rev Cancer. 2013;13(10):701–713. doi:10.1038/nrc3582
12. Hu CC, Liang YW, Hu JL, Liu LF, Liang JW, Wang R. LncRNA RUSC1-AS1 promotes the proliferation of breast cancer cells by epigenetic silence of KLF2 and CDKN1A. Eur Rev Med Pharmacol Sci. 2019;23(15):6602–6611. doi:10.26355/eurrev_201908_18548
13. Lu F, Li C, Yu Y, et al. [KLF3 regulates the movement, migration and invasion of breast cancer cells through STAT3]. Zhonghua Yi Xue Za Zhi. 2019;99(38):3014–3018. Chinese. doi:10.3760/cma.j.issn.0376-2491.2019.38.010
14. Hu D, Zhou Z, Davidson NE, Huang Y, Wan Y. Novel insight into KLF4 proteolytic regulation in estrogen receptor signaling and breast carcinogenesis. J Biol Chem. 2012;287(17):13584–13597. doi:10.1074/jbc.M112.343566
15. Chen CH, Yang N, Zhang Y, et al. Inhibition of super enhancer downregulates the expression of KLF5 in basal-like breast cancers. Int J Biol Sci. 2019;15(8):1733–1742. doi:10.7150/ijbs.35138
16. Guo P, Dong XY, Zhao KW, Sun X, Li Q, Dong JT. Estrogen-induced interaction between KLF5 and estrogen receptor (ER) suppresses the function of ER in ER-positive breast cancer cells. Int J Cancer. 2010;126(1):81–89. doi:10.1002/ijc.24696
17. Liu J, Du T, Yuan Y, He Y, Tan Z, Liu Z. KLF6 inhibits estrogen receptor-mediated cell growth in breast cancer via a c-Src-mediated pathway. Mol Cell Biochem. 2010;335(1–2):29–35. doi:10.1007/s11010-009-0237-8
18. Hatami R, Sieuwerts AM, Izadmehr S, et al. An oncogenic splice variant drives EMT and metastasis in breastcancer. Cancer Discov. 2013;3(3):Of16. doi:10.1158/2159-8290.CDRW2013-027
19. Wang X, Lu H, Urvalek AM, et al. KLF8 promotes human breast cancer cell invasion and metastasis by transcriptional activation of MMP9. Oncogene. 2011;30(16):1901–1911. doi:10.1038/onc.2010.563
20. Lu H, Hu L, Yu L, et al. KLF8 and FAK cooperatively enrich the active MMP14 on the cell surface required for the metastatic progression of breast cancer. Oncogene. 2014;33(22):2909–2917. doi:10.1038/onc.2013.247
21. Bai XY, Li S, Wang M, et al. Kruppel-like factor 9 down-regulates matrix metalloproteinase 9 transcription and suppresses human breast cancer invasion. Cancer Lett. 2018;412:224–235. doi:10.1016/j.canlet.2017.10.027
22. Gumireddy K, Li A, Gimotty PA, et al. KLF17 is a negative regulator of epithelial-mesenchymal transition and metastasis in breast cancer. Nat Cell Biol. 2009;11(11):1297–1304. doi:10.1038/ncb1974
23. Hsu CF, Sui CL, Wu WC, et al. Klf10 induces cell apoptosis through modulation of BI-1 expression and Ca2+ homeostasis in estrogen-responding adenocarcinoma cells. Int J Biochem Cell Biol. 2011;43(4):666–673. doi:10.1016/j.biocel.2011.01.010
24. Jin W, Chen BB, Li JY, et al. TIEG1 inhibits breast cancer invasion and metastasis by inhibition of epidermal growth factor receptor (EGFR) transcription and the EGFR signaling pathway. Mol Cell Biol. 2012;32(1):50–63. doi:10.1128/MCB.06152-11
25. Han M, Wang Y, Guo G, et al. microRNA-30d mediated breast cancer invasion, migration, and EMT by targeting KLF11 and activating STAT3 pathway. J Cell Biochem. 2018;119(10):8138–8145. doi:10.1002/jcb.26767
26. Guan B, Li Q, Shen L, et al. MicroRNA-205 directly targets Krüppel-like factor 12 and is involved in invasion and apoptosis in basal-like breast carcinoma. Int J Oncol. 2016;49(2):720–734. doi:10.3892/ijo.2016.3573
27. Fan G, Sun L, Shan P, et al. Loss of KLF14 triggers centrosome amplification and tumorigenesis. Nat Commun. 2015;6:8450. doi:10.1038/ncomms9450
28. Yoda T, McNamara KM, Miki Y, et al. KLF15 in breast cancer: a novel tumor suppressor?. Cell Oncol. 2015;38(3):227–235.
29. Kim HS, Lee JU, Yoo TK, et al. Omission of chemotherapy for the treatment of mucinous breast cancer: a nationwide study from the Korean Breast Cancer Society. J Breast Cancer. 2019;22(4):599–612. doi:10.4048/jbc.2019.22.e46
30. Yan Q, Zhang W, Wu Y, et al. KLF8 promotes tumorigenesis, invasion and metastasis of colorectal cancer cells by transcriptional activation of FHL2. Oncotarget. 2015;6(28):25402–25417. doi:10.18632/oncotarget.4517
31. Suh Y, Noh SJ, Lee S, et al. Dopamine D1 receptor (D1R) expression is controlled by a transcriptional repressor complex containing DISC1. Mol Neurobiol. 2019;56(10):6725–6735.
32. Jang MK, Lee S, Jung MH. RNA-Seq analysis reveals a negative role of KLF16 in adipogenesis. PLoS One. 2016;11(9):e0162238. doi:10.1371/journal.pone.0162238
33. Daftary GS, Lomberk GA, Buttar NS, et al. Detailed structural-functional analysis of the Kruppel-like factor 16 (KLF16) transcription factor reveals novel mechanisms for silencing Sp/KLF sites involved in metabolism and endocrinology. J Biol Chem. 2012;287(10):7010–7025. doi:10.1074/jbc.M111.266007
34. Chen X, Li S, Ke Y, et al. KLF16 suppresses human glioma cell proliferation and tumourigenicity by targeting TFAM. Artif Cells Nanomed Biotechnol. 2018;46(sup1):608–615. doi:10.1080/21691401.2018.1431654
35. Ma P, Sun CQ, Wang YF, et al. KLF16 promotes proliferation in gastric cancer cells via regulating p21 and CDK4. Am J Transl Res. 2017;9(6):3027–3036.
36. Xiong Y, Wang Z, Zhou Q, et al. Identification and validation of dichotomous immune subtypes based on intratumoral immune cells infiltration in clear cell renal cell carcinoma patients. J Immunother Cancer. 2020;8(1):e000447. doi:10.1136/jitc-2019-000447
37. Weinberger P, Ponny SR, Xu H, et al. Cell cycle M-Phase genes are highly upregulated in anaplastic thyroid carcinoma. Thyroid. 2017;27(2):236–252. doi:10.1089/thy.2016.0285
38. Haffty BG, Yang Q, Reiss M, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24(36):5652–5657. doi:10.1200/JCO.2006.06.5664
39. Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26(8):1275–1281. doi:10.1200/JCO.2007.14.4147
40. Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7(344):re8. doi:10.1126/scisignal.2005189
41. Xia E, Bhandari A, Shen Y, Zhou X, Wang O. lncRNA LINC00673 induces proliferation, metastasis and epithelial-mesenchymal transition in thyroid carcinoma via Kruppel-like factor 2. Int J Oncol. 2018;53(5):1927–1938. doi:10.3892/ijo.2018.4524
42. Qin J, Zhou Z, Chen W, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471. doi:10.1038/ncomms9471
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.