Back to Journals » International Journal of General Medicine » Volume 15
The Association of TNF-α Promoter Polymorphisms with Genetic Susceptibility to Cervical Cancer in a Chinese Han Population
Authors Yang J, Wang Y, Zhang S, Li Y, Li C, Liu W, Liu S, Liang Y, Zhang X, Yan Z, Shi L, Yao Y
Received 19 November 2021
Accepted for publication 23 December 2021
Published 10 January 2022 Volume 2022:15 Pages 417—427
DOI https://doi.org/10.2147/IJGM.S350263
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Jia Yang,1,* Yingying Wang,2,* Shao Zhang,3 Yu Li,4 Chuanyin Li,1 Weipeng Liu,1 Shuyuan Liu,1 Yan Liang,1 Xinwen Zhang,1 Zhiling Yan,3 Li Shi,1 Yufeng Yao1
1Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Kunming, Yunnan, 650118, People’s Republic of China; 2School of Basic Medical Science, Kunming Medical University, Kunming, 650500, People’s Republic of China; 3Department of Gynaecologic Oncology, The No. 3 Affiliated Hospital of Kunming Medical University, Kunming, 650118, People’s Republic of China; 4Department of Obstetrics, The No. 1 People’s Hospital of Kunming, Kunming, 650011, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yufeng Yao
Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Kunming, 650118, Yunnan, People’s Republic of China
Email [email protected]; [email protected]
Background: The tumour necrosis factor-α (TNF-α) gene plays an important role in the host immune response, which will influence the development and clearance of cancer. Polymorphism of the TNF-α promoter region is considered to influence its transcription and be a risk factor for tumorigenesis. In the current study, we evaluated the role of TNF-α promoter region polymorphisms in susceptibility to cervical intraepithelial neoplasia (CIN) and cervical cancer (CC).
Methods: A total of 2732 subjects, including 1173 healthy controls, 579 patients with CIN and 980 patients with CC in a Chinese Han population, were selected for the current study. Five SNPs in the TNF-α promoter, rs1799964 (− 1031 C>T), rs1800630 (− 863 A>C), rs1799724 (− 857 C>T), rs1800629 (− 308 A>G) and rs361525 (− 238 A>G), were selected and genotyped using TaqMan Assays. The association of these SNPs with CIN and cervical cancer was evaluated among healthy controls, CIN and CC patients.
Results: The frequency distribution of rs1800629 and rs361525 alleles was significantly different between the CC group and the control group (P=0.009 and P=0.002). The rs1800629 A allele was found to be a protective factor for CC (OR=0.72; 95% CI=0.56– 0.92). The rs361525 A allele was found to be a risk factor for CC (OR=1.69; 95% CI=1.21– 2.38). After pathological subtyping of CC, the allele distribution of rs1800629 and rs361525 were both significantly different between the cervical squamous cell carcinoma and control groups (P=0.002 and P< 0.001). The rs1800629 A allele was protective factor for cervical squamous cell carcinoma (OR=0.66; 95% CI=0.50– 0.86). The rs361525 A allele was a risk factor for cervical squamous cell carcinoma (OR=1.87; 95% CI=1.32– 2.65). Moreover, the genotypic frequency of rs361525 was significantly different between cervical cancer stage I and stage II (P=0.003).
Conclusion: The rs1800629 and rs361525 in the TNF-α promoter are associated with susceptibility to CC in the Chinese Han population.
Keywords: cervical intraepithelial neoplasia, CIN, cervical cancer, CC, promoter, single-nucleotide polymorphism, SNP, tumour necrosis factor-α, TNF-α
Introduction
Cervical cancer is the fourth most common cancer among women worldwide and the second leading cause of cancer death in developing countries.1 Persistent infection with high-risk human papilloma virus is considered to be the main cause of cervical cancer. Epidemiological studies have shown that most cases of HPV infection can be cleared by the host immune system, with only a small portion of chronically infected patients and further developing cervical intraepithelial neoplasia (CIN) and progressing to cervical cancer.2
The host immune system plays an important role in the clearance and development of cancers. As one of immune system proinflammatory cytokine, tumour necrosis factor (TNF), which includes tumour necrosis factor-α (TNF-α) and tumour necrosis factor-β (TNF-β), participates in multiple biological activities. TNF-α, which is mainly produced by monocytes or macrophages, is a pro-inflammatory cytokine that plays an important role in homeostasis of the immune system and host defense.3,4 TNF-α is highly expressed in a variety of tumours and participates in cell transformation.5 Overall, TNF-α promotes tumour cell proliferation, invasion and metastasis by regulating tumour angiogenesis.
The human TNF-α gene is 2.76 kb long and located on chromosome 6; it contains 3 introns and 4 exons and is closely related to the major histocompatibility complex region. TNF-α polymorphisms are mainly concentrated in the promoter region and closely related to the risk of cervical cancer. Nevertheless, the results of associations between TNF-α polymorphisms and cervical cancer are still controversial in different populations, even the same population.6–15 For example, in 2001, Jang et al reported that the rs361525 A allele is a protective factor for cervical cancer in a Korean population.13 In 2018, Li et al reported that the rs361525 A allele is a risk factor for cervical squamous cell carcinoma in a Chinese population from Shandong Province, Northern China.14 For another example, Du et al also found the rs1800629 A allele to be associated with an increased risk of cervical cancer in a Chinese population from Sichuan Province, Southwest of China.7 However, in 2012, Wang et al reported that rs1800629 was not significantly associated with cancer in a Chinese population from Liaoning Province, Northeast of China.10 Thus, more populations from different regions should be investigated.
In the current study, five single-nucleotide polymorphisms (SNPs) located in the TNF-α promoter, rs1799964 (−1031 C>T), rs1800630 (−863 A>C), rs1799724 (−857 C>T), rs1800629 (−308 A>G) and rs361525 (−238 A>G), were selected to investigate allele distribution and genotypes in CIN, cervical cancer and healthy controls in a Chinese Han population to clarify the role of these SNPs in the occurrence and development of cervical cancer.
Materials and Methods
Ethics Declarations
The current study obtained approval from the Institutional Review Board of the No. 3 Affiliated Hospital of Kunming Medical University. The protocol employed in this investigation was in accordance with the principles expressed in the Helsinki Declaration of 1975, as revised in 2008.
Subjects
A total of 2732 subjects, including 1173 healthy controls, 579 patients with CIN, and 980 patients with cervical cancer (CC), were recruited to participate in the present study. The CIN and cervical cancer patients were all diagnosed at the Third Affiliated Hospital of Kunming Medical University from October 2013 to May 2018. Inclusion criteria for the patients were as follows: (1) CIN or cervical cancer diagnosed according to the World Health Organization Comprehensive Cervical Cancer Control: A Guide to Essential Practice16 and the International Federation of Gynaecology and Obstetrics, 2009; (2) no other malignancy; and (3) no preoperative neoadjuvant therapy (including chemotherapy and radiotherapy). The exclusion criteria for the patients were as follows: (1) presented with a history of primary cancer other than cervical cancer; (2) malignant tumours other than cervical cancer; and (3) receiving radiotherapy or chemotherapy with unclear pathological diagnosis. Over the same period, 1173 women from a healthy screening at the same hospital served as healthy controls in the present study. The inclusion criteria for control individuals were as follows: (1) absence of any malignancy history; (2) absence of any cervical lesion; (3) tested negative for HPV; and (4) no chronic diseases. All subjects were Han Chinese from Yunnan Province (Southwest of China) and signed informed consent. The geographic origin and pedigree (unrelated through at least three generations) of each individual were ascertained.
DNA Extraction
A total of 5 mL of fasting venous blood was collected from the subjects. Genomic DNA from peripheral blood was extracted using a whole-blood genomic DNA mini kit to extract DNA (QIAamp DNA Blood Mini Kit). An ultramicro UV-visible spectrophotometer (ND-2000, Thermo Scientific, USA) was used to detect the concentration and purity of the DNA.
SNP Genotyping
The probes and primers used for genotyping these five SNPs were all purchased from ABI (http://www.appliedbiosystems.com). Primers and probes for the genotyping were commercially available. The array ID were C___7514871_10(rs1799964), C__11918223_10(rs179972), C___7514879_10(rs1800629), C___2215707_10(rs361525). For rs1800630, the primers and probes for the genotyping were not commercially available. Thus, the primers and probe were designed by ABI. The primer sequence is: forward 5ʹ-GGGCTATGGAAGTCGAGTATGG-3ʹ, reverse 5ʹ-CCCTCTACATGGCCCTGTCT-3ʹ; VIC probe sequence is ACCCCCACTTAACG, FAM probe sequence is ACCCCCCCTTAACG. The five SNPs were genotyped using the TaqMan fluorescent quantitative PCR method with a QuantStudio™ Real-Time PCR instrument. The TaqMan Genotyping Master Mix used in the typing test was purchased from ABI. The PCR volume was 5 µL, and the reaction conditions were 95°C predenaturation for 10 minutes, 40 cycles of 95°C denaturation for 15 seconds and 60°C annealing for 1 minute, and 60°C extension for 5 minutes. Deionized water was used to replace the template DNA as a negative control. The PCR experiment data were analysed using TaqMan Genotyper Software (Version 1.3.1). To identify the accuracy of SNP genotyping using the TaqMan assay, samples of each genotype of the five SNPs were sequenced. Then, each genotype samples were used as positive sample in TaqMan assay
Statistical Analysis
Hardy–Weinberg equilibrium (HWE) was calculated using Plink software. Differences in age among the CIN group, cancer group and control group were analysed using one-way ANOVA (Analysis of Variance) with GraphPad Prism 7 software. The associations between the five SNP alleles and genotypes and the CIN, CC groups were analysed using the SHEsis program.17,18 The association of the genotypes of these SNPs with CIN and CC was evaluated using inheritance model analysis with SNPstats software.19 The analysed inheritance models included the codominant model, dominant model, recessive model, overdominant model and log-additive model. The Akaike information criterion (AIC) and the Bayesian information criterion (BIC) were applied to determine the best fit model for each SNP.19 The inheritance model corresponding to the smallest AIC and BIC was the best fit model. Bonferroni correction was applied in multiple comparisons. A difference was considered statistically significant at P<0.01 (0.05/5).
Results
Subject Characteristics
Table 1 shows the clinical data for the subjects in this study. The cervical cancer group contained 980 patients, including 794 cases of squamous cell carcinoma (SCC), 162 cases of adenocarcinoma (AC), and 24 cases of other types of cancer. According to cervical cancer staging, 679 patients were in stage I, 265 in stage II, and 36 in stage III+IV. The CIN group contained 579 cases, of which 120 were in CIN stage I, 104 in CIN stage II, and 355 in CIN stage III. Comparing the ages among the control, CIN and CC groups, no statistically significant difference among the three groups was observed (F=2.082, P=0.125).
 |
Table 1 Characteristics of the Subjects Enrolled in the Current Study |
Association Analysis of Five SNPs in the TNF-α Promoter with CIN and Cervical Cancer
The alleles and genotype frequencies of the five SNPs in the TNF-α promoter among the control, CIN and cervical cancer groups are shown in Table 2. The results of HWE showed that rs361525, rs1799964 and rs1800629 were in HWE except for the rs1799724 and rs1800630 in the control group (P<0.05). For the control and CC groups, the allele frequencies of rs1800629 were significantly different between these two groups after Bonferroni correction (P=0.009), which indicates that the A allele is a protective factor against cervical cancer (OR=0.72; 95% CI=0.56–0.92). For rs361525, the allele and genotype frequencies were significantly different between these two groups after Bonferroni correction (P=0.002 and P=0.009, respectively), and the A allele might be a risk factor for cervical cancer (OR=1.69; 95% CI=1.21–2.38). For the control and CIN groups, no difference of alleles and genotypes was observed between these two groups after Bonferroni correction (P>0.01).
 |
Table 2 The Allelic and Genotypic Distribution of Five SNPs in TNF-α Gene Among Control, CIN and Cervical Cancer Groups |
Inheritance Model Analysis of Five SNPs in the TNF-α Promoter in CIN and Cervical Cancer
The results of inheritance model analysis of five SNPs in the TNF-α promoter among the control, CIN and cervical cancer groups are shown in Table 3. Comparison between the cervical cancer and control groups showed that the dominant inheritance model was the best fit model for rs1800630. In this model, the A/C-A/A genotype was related to a reduced risk of cervical cancer (P=0.006, OR=0.77; 95% CI=0.64–0.93). For rs1800629, the overdominant inheritance model was the best fit. In this model, the A/G genotype was related to a reduced risk of cervical cancer (P=0.008, OR=0.70; 95% CI=0.53–0.91). For rs361525, the dominant inheritance model was the best fit, in which the A/G-A/A genotype was related to an increased risk of cervical cancer (P=0.008, OR=1.63; 95% CI=1.14–2.35). Comparison between the CIN and control groups showed that no significant differences between the control group and the CIN group in the inheritance models (P>0.01).
 |
Table 3 Inheritance Model Analysis of Five SNPs in TNF-α Gene Among Control, CIN and Cervical Cancer Groups |
Association Analysis of Five SNPs in the TNF-α Promoter with Different Pathological Types of Cervical Cancer
Comparison between patients with different pathological types of cervical cancer and healthy controls is shown in Table 4. The allelic frequency and genotype frequency of rs1800629 were significantly different between the SCC group and the control group (P=0.002 and P=0.006, respectively), which indicates that the A allele of this SNP is a protective factor for SCC (OR=0.66; 95% CI=0.50–0.86). The frequency of rs361525 was also significantly different between the SCC group and the control group (P<0.001 and P=0.002 respectively), and the A allele was a risk factor for SCC (OR=1.87; 95% CI=1.32–2.65). In addition, the frequencies of the five SNPs showed no significant differences between the AC group and the control group (P>0.01).
 |
Table 4 The Allelic and Genotypic Distribution of Five SNPs in TNF-α Gene Among Control, Cervical Squamous Cell Carcinoma and Adenocarcinoma |
Inheritance Model Analysis of Five SNPs in the TNF-α Promoter with Different Pathological Types of Cervical Cancer
The results of analysis of different pathological types of cervical cancer under different inheritance models are shown in Table 5. For rs1800629, comparison between the SCC group and the control group showed that the overdominant model was the best fit. In this model, the A/G genotype was related to a reduced risk of SCC (P=0.002, OR=0.63; 95% CI=0.47–0.84). For rs361525, the dominant and log-additive inheritance models were best fitting models. The A/G-A/A genotype was related to an increased risk of SCC in the dominant model (P=0.002, OR=1.80; 95% CI=1.24–2.61) and 2A/A-A/G genotype was related to an increased risk of SCC in the log-additive inheritance model (P=0.002, OR=1.74; 95% CI=1.22–2.48). No significant differences between the AC and control group were found for the frequencies of the five SNPs in the inheritance model (P>0.01).
 |
Table 5 Inheritance Model Analysis of Five SNPs in TNF-α Gene Among Control, Cervical Squamous Cell Carcinoma and Adenocarcinoma |
Association Analysis of Five SNPs in the TNF-α Promoter with Different Stages of Cervical Cancer
As only 36 patients were stage III+IV of cervical cancer, we just analysed the association of five SNPs in the TNF-α promoter with stage I and stage II of cervical cancer. Comparison between stage I (n=679) and stage II (n=265) of cervical cancer is shown in Table 6. The genotypic frequency of rs361525 was significantly different between stage I and stage II (P=0.003). The allelic frequency showed no significantly difference. In addition, the frequencies of the other four SNPs showed no significant differences between stage I and stage II (P>0.01).
 |
Table 6 The Allelic and Genotypic Distribution of SNPs in TNF-α Gene Promoter Between Stage I and Stage II of Cervical Cancer |
Inheritance Model Analysis of Five SNPs in the TNF-α Promoter with Different Stages of Cervical Cancer
The results of analysis of different stage I and stage II of cervical cancer under different inheritance models are shown in Table 7. For rs361525, the frequency of A/A genotype was 0% and 1.1% in stage I and stage II, respectively. The recessive inheritance models were best fitting models and the A/A genotype was related to an increased risk of stage in recessive inheritance model (P=0.004). No significant differences between stage I and stage II were found for the frequencies of the other four SNPs in the inheritance model (P>0.01).
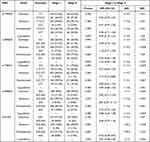 |
Table 7 Inheritance Model Analysis of SNPs in TNF-α Gene Promoter Between Stage I and Stage II of Cervical Cancer |
Discussion
Cervical cancer is a gynaecological malignancy with a high incidence worldwide that seriously influences women’s lives. Many studies have shown that host genetic variation, especially immune genes variations, has a certain association with susceptibility to cervical cancer.20–23 In the current study, we found that rs1800629 and rs361525 in the TNF-α gene promoter are related to susceptibility to cervical cancer in a Chinese Han population.
Recently, a Meta-Analysis found that rs1800629 (−308 A>G) was associated with the cervical cancer in different inheritance models in the general population.24 In 2005, Duarte et al reported that the rs1800629 A allele is associated with an increased risk of invasive cervical cancer in the Portuguese population.6 Then, Singh et al and Du et al also found the rs1800629 A allele to be associated with an increased risk of cervical cancer in an Indian population and a Chinese population from Sichuan Province, Southwest of China, respectively.7,8 However, we found that the rs1800629 A allele is a protective factor for cervical cancer, and the A/G genotype was related to a reduced risk of cervical cancer in the current study. Our findings are similar to those of Zidi et al, which also found that A/G genotype is a protective factor for cervical cancer in Tunisia.11 The discrepancy between Du and Singh et al study and our study could be due to the relatively small sample sizes and population genetic background. For example, the sample sizes in Du et al and Singh et al involved 522 and 150 cervical cancer patients, 550 and 162 healthy individuals, whereas 1173 healthy controls, 579 patients with CIN, and 980 patients with cervical cancer were enrolled in the current study. In 1997, Kroeger et al demonstrated that rs1800629 affected the affinity of factor binding and resulted in a factor binding to A allele but not G allele.25 Their results showed that the level of transcription of rs1800629 A allele has increased twofold greater level by comparing to that of the G allele. In 2019, Du et al found the PBMCs carrying the rs1800629 AA genotype showed significantly higher rates of T-cell proliferation which play a major role in the surveillance of cancer cells. Thus, the rs1800629 AA genotype positively regulate T-cell activation, and might confer reduced susceptibility to cervical cancer.7 Thus, the rs1800629 A allele and AA genotype could increase the TNF-α level and T-cell activation to protect from cervical cancer.
Recently, Wang et al reported that rs361525 was associated with the cervical cancer in Asians using meta-analysis.24 In 2019, Du et al showed that rs361525 A allele and AA genotype is a protective factor against cervical cancer in Sichuan Province, Southwest of China.7 In 2018, Li et al reported that the rs361525 A allele is a risk factor for SCC in a Chinese population from Shandong Province, North of China.14 In the current study, we also found the rs361525 A allele and A/G-A/A genotype to be a risk factor for cervical cancer in a Chinese population, which was similar to the meta-analysis results of de Moura et al, which showed that rs361525 A/A genotype in codominant model and recessive model with increased risk for cervical cancer.26 In the de Moura study, they also identified two transcription factors, MZF1 and ZNF263, which binds to rs1800629 and rs361525 in the TNF-α promote by using bioinformatic method. The binding could affect the affinity between promoter and transcription factors and influence the expression of TNF-α transcriptional levels, which is associated with cervical cancer.26 In addition, in subgroup analysis, the A allele was a risk factor for SCC but not AC. Our results are similar to those of Li et al, in which the rs361525 A allele was a risk factor for cervical cancer, especially SCC. One of the reasons for the discrepancy between the studies of Du et al and ours might be the different pathological types of cervical cancer included. We found that the A allele of rs361525 is a risk factor for SCC, which indicates that the A allele has different roles in pathological types of cervical cancer. SCC is the most common pathological type of cervical cancer, accounting for approximately 75% to 80%, followed by AC, which accounts for approximately 10.0% to 25%.27 Therefore, it is necessary to study the role of rs361525 in the different pathological types of cervical cancer in the future.
In the current study, we found that rs1800629 and rs361525 in the TNF-α promoter gene are associated with cervical cancer and SCC in a Chinese Han population. The A allele of rs1800629 is a protective factor for cervical cancer and SCC, whereas the A allele of rs361525 is a risk factor for cervical cancer and SCC. Nevertheless, association results vary among different populations in different studies. As a result, multicentre and more samples from different regions should be evaluated to study the association between TNF-α gene polymorphisms and cervical cancer. Moreover, the function of polymorphisms should be investigated in the future.
Abbreviations
TNF-α, tumour necrosis factor-α; TNF-β, tumour necrosis factor-β; CIN, cervical intraepithelial neoplasia; CC, cervical cancer SNPs, single nucleotide polymorphisms; ANOVA, analysis of variance; HWE, Hardy–Weinberg equilibrium; ORs, odds ratios; CIs, confidence intervals: AIC, Akaike information criterion; BIC, Bayesian information criterion; SCC, squamous cell carcinoma; AC, adenocarcinoma.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
The current study was supported by grants from the PUMC Youth Fund (3332019111); National Science Foundation of China (82103190); Yunnan Province Clinical Research Center for Gynecological and Obstetric Disease (2022YJZX-FC05); Special Funds for High-level Healthy Talents of Yunnan Province (L-201615, H-2018014 and D-2018037). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Disclosure
The authors declare that they have no competing interests.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Shanmugasundaram S, You J. Targeting persistent human papillomavirus infection. Viruses. 2017;9(8):229. doi:10.3390/v9080229
3. Balkwill F. Tumor necrosis factor or tumor promoting factor? Cytokine Growth Factor Rev. 2002;13(2):135–141. doi:10.1016/S1359-6101(01)00020-X
4. Balkwill F. TNF-alpha in promotion and progression of cancer. Cancer Metastasis Rev. 2006;25(3):409–416. doi:10.1007/s10555-006-9005-3
5. Komori A, Yatsunami J, Suganuma M, et al. Tumor necrosis factor acts as a tumor promoter in BALB/3T3 cell transformation. Cancer Res. 1993;53(9):1982–1985.
6. Duarte I, Santos A, Sousa H, et al. G-308A TNF-alpha polymorphism is associated with an increased risk of invasive cervical cancer. Biochem Biophys Res Commun. 2005;334(2):588–592. doi:10.1016/j.bbrc.2005.06.137
7. Du GH, Wang JK, Richards JR, Wang JJ. Genetic polymorphisms in tumor necrosis factor alpha and interleukin-10 are associated with an increased risk of cervical cancer. Int Immunopharmacol. 2019;66:154–161. doi:10.1016/j.intimp.2018.11.015
8. Singh H, Jain M, Sachan R, Mittal B. Association of TNFA (−308G>A) and IL-10 (−819C>T) promoter polymorphisms with risk of cervical cancer. Int J Gynecol Cancer. 2009;19(7):1190–1194. doi:10.1111/IGC.0b013e3181a3a3af
9. Behboodi N, Farazestanian M, Rastgar-Moghadam A, et al. Association of a variant in the tumor necrosis factor alpha gene with risk of cervical cancer. Mol Biol Rep. 2021;48(2):1433–1437. doi:10.1007/s11033-021-06185-4
10. Wang N, Yin D, Zhang S, et al. TNF-alpha rs1800629 polymorphism is not associated with HPV infection or cervical cancer in the Chinese population. PLoS One. 2012;7(9):e45246. doi:10.1371/journal.pone.0045246
11. Zidi S, Stayoussef M, Zouidi F, et al. Tumor necrosis factor alpha (−238/-308) and TNFRII-VNTR (−322) polymorphisms as genetic biomarkers of susceptibility to develop cervical cancer among tunisians. Pathol Oncol Res. 2015;21(2):339–345. doi:10.1007/s12253-014-9826-2
12. Babapour N, Mehramiz M, Rastgar Moghadam A, et al. Association of TNF-308 G>A polymorphism located in tumor necrosis factor a with the risk of developing cervical cancer and results of pap smear. J Cell Biochem. 2019;120(4):5444–5448. doi:10.1002/jcb.27823
13. Jang WH, Yang YI, Yea SS, et al. The −238 tumor necrosis factor-alpha promoter polymorphism is associated with decreased susceptibility to cancers. Cancer Lett. 2001;166(1):41–46. doi:10.1016/S0304-3835(01)00438-4
14. Li X, Yin G, Li J, et al. The correlation between tnf-α promoter gene polymorphism and genetic susceptibility to cervical cancer. Technol Cancer Res Treat. 2018;17:1533033818782793. doi:10.1177/1533033818782793
15. Duvlis S, Dabeski D, Cvetkovski A, Mladenovska K, Plaseska-Karanfilska D. Association of TNF-A (rs361525 and rs1800629) with susceptibility to cervical intraepithelial lesion and cervical carcinoma in women from Republic of North Macedonia. Int J Immunogenet. 2020;47(6):522–528. doi:10.1111/iji.12506
16. World Health Organization. WHO guidelines on malaria approved by the guidelines review committee; 2013.
17. Li Z, Zhang Z, He Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the shesis. Cell Res. 2009;19(4):519–523. doi:10.1038/cr.2009.33
18. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–98. doi:10.1038/sj.cr.7290272
19. Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi:10.1093/bioinformatics/btl268
20. Calhoun ES, McGovern RM, Janney CA, et al. Host genetic polymorphism analysis in cervical cancer. Clin Chem. 2002;48(8):1218–1224. doi:10.1093/clinchem/48.8.1218
21. Hildesheim A, Wang SS. Host and viral genetics and risk of cervical cancer: a review. Virus Res. 2002;89(2):229–240. doi:10.1016/S0168-1702(02)00191-0
22. Madeleine MM, Johnson LG, Smith AG, et al. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res. 2008;68(9):3532–3539. doi:10.1158/0008-5472.CAN-07-6471
23. Yang J, Liu W, Yan Z, et al. Polymorphisms in transporter associated with antigen presenting are associated with cervical intraepithelial neoplasia and cervical cancer in a Chinese Han population. HLA. 2021;98(1):23–36. doi:10.1111/tan.14333
24. Wang Y, Yang J, Huang J, Tian Z. Tumor necrosis factor-alpha polymorphisms and cervical cancer: evidence from a meta-analysis. Gynecol Obstet Invest. 2020;85(2):153–158. doi:10.1159/000502955
25. Kroeger KM, Carville KS, Abraham LJ. The −308 tumor necrosis factor-alpha promoter polymorphism effects transcription. Mol Immunol. 1997;34(5):391–399. doi:10.1016/S0161-5890(97)00052-7
26. de Moura EL, Dos santos ACM, da Silva DM, et al. Association of polymorphisms in cytokine genes with susceptibility to precancerous lesions and cervical cancer: a systematic review with meta-analysis. Immunol Invest. 2021;50(5):492–526. doi:10.1080/08820139.2020.1778023
27. Gadducci A, Guerrieri ME, Cosio S. Adenocarcinoma of the uterine cervix: pathologic features, treatment options, clinical outcome and prognostic variables. Crit Rev Oncol Hematol. 2019;135:103–114. doi:10.1016/j.critrevonc.2019.01.006
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
