Back to Journals » OncoTargets and Therapy » Volume 9
The association of PTEN hypermethylation and breast cancer: a meta-analysis
Received 29 April 2016
Accepted for publication 6 July 2016
Published 12 September 2016 Volume 2016:9 Pages 5643—5650
DOI https://doi.org/10.2147/OTT.S111684
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Min Li
Shanshan Luo, Jiansi Chen, Xianwei Mo
Department of Gastrointestinal Surgery, Tumor Hospital of Guangxi Medical University, Nanning, Guangxi, People’s Republic of China
Objective: Phosphatase and tensin homolog (PTEN) deleted on chromosome 10, as a tumor suppressor gene, is crucial for the development of both familial and sporadic breast cancer (BC). The aim of this study was to perform a meta-analysis to evaluate the clinicopathological significance of PTEN promoter hypermethylation in BC.
Methods: A comprehensive literature search was made in PubMed, Embase, Google Scholar, Chinese database (China National Knowledge Infrastructure [CNKI]), and Web of Science. The analysis of pooled data was performed with Review Manager 5.2. The fixed-effects or random-effects models were used to evaluate odds ratios (ORs) and 95% confidence intervals (CIs).
Results: The meta-analysis included eight studies and a total of 923 patients. The frequency of PTEN promoter hypermethylation was significantly increased in ductal carcinoma in situ (DCIS) and invasive ductal carcinoma (IDC) compared to normal breast tissues (OR =22.53, P=0.0002 and OR =22.86, P<0.00001, respectively). However, the frequency of PTEN promoter hypermethylation was similar between IDC and DCIS. Additionally, PTEN methylation was not significantly correlated to estrogen receptor (ER) or human epidermal growth factor type 2 (HER-2) status in patients with BC.
Conclusion: PTEN promoter hypermethylation is significantly associated with the risk of DCIS and IDC, suggesting PTEN promoter hypermethylation is a valuable biomarker for diagnosis of BC.
Keywords: breast cancer, PTEN, meta-analysis, methylation, estrogen receptor, HER-2
Introduction
Breast cancer (BC) is the most common malignant disease in women worldwide, and the principal cause of cancer-related female mortality globally.1 The development of BC involves a series of steps: initiating atypical ductal hyperplasia (ADH), followed by subsequent evolution to ductal carcinoma in situ (DCIS), culminating as invasive ductal carcinoma (IDC), and finally advancing to metastatic disease.2 Molecular studies have demonstrated the great heterogeneity of IDC. Based on gene expression profiling and epithelial cell types, IDC is divided into four molecular subtypes: estrogen receptor (ER)-positive luminal A, ER-positive luminal B, ER-negative/human epidermal growth factor type 2 (HER-2), and basal subtype. Luminal A and B are most common subtypes, representing low- or intermediate-grade tumors. HER-2 and basal subtypes represent high-grade tumors and are characterized by lack of expression of ER and progesterone receptor (PR), displaying necrosis and lymphocytic infiltrate as well as poor response to treatment.3 Given the variability in the clinical progression of disease, it becomes critical to identify markers for early detection of BC and prediction of tumor behavior.
Breast carcinogenesis is a stepwise accumulation of genetic changes including point mutations, deletions, oncogene activation, or tumor suppressor inactivation.4 Epigenetics refers to heritable changes in gene expression that do not involve changes to the underlying DNA sequence and has received significantly more attention from researchers over the last decade.5 Epigenetic alterations occur in malignant transformation which involves changes in DNA methylation, including global hypomethylation, focal hypermethylation, histone modifications, and nucleosomal remodeling.6 Phosphatase and tensin homolog (PTEN) was deleted in breast cancer on chromosome 10, located on chromosomal subband 10q23.3, and is a tumor suppressor which is frequently mutated in human cancers.7 PTEN regulates a variety of essential processes such as translation, cell cycle progression, and apoptosis by blocking the activation of the serine/threonine kinase Akt/PKB. Reduced or lost PTEN expression was observed in many human tumors, including brain tumors,8 melanomas,9 hepatocellular carcinomas,10 thyroid carcinomas,11 endometrial carcinomas,12 lymphoid neoplasia,13 and breast carcinomas.14–17 Bose et al observed that the expression of PTEN protein is reduced or lost in 38% of invasive BCs.14 Moreover, numerous studies have demonstrated that methylation is the mechanism for PTEN inactivation in BC.18–21 The inactivation of PTEN has been considered to be caused by hypermethylation of its promoter in BC; the reported rates of PTEN promoter hypermethylation in BC are notably diverse. In addition, whether or not the inactivation is associated with the incidence and clinicopathological significance have not been thoroughly examined. The heterogeneous results of these studies do need further evaluation of the relationship between the promoter hypermethylation status of PTEN and BC. In this study, we quantified the association between PTEN promoter hypermethylation and BC using meta-analysis methods.
Methods
Selection criteria and study search
We comprehensively reviewed electronic databases including PubMed (1966 to January 2016), Web of Science (1945 to January 2016), Embase (1980 to January 2016), Cochrane Library Database (1972 to January 2016), Google Scholar, and Chinese database CNKI. The keywords “PTEN methylation” and “breast cancer” or “breast carcinoma” were used for relative studies searching without any language restriction. There were 48 studies identified from PubMed, 120 studies from Web of Science, 97 studies from Embase, and 350 studies from Google Scholar. A total of 615 studies were reviewed by titles and abstracts. We manually searched potentially relevant articles from the reference lists of retrieved articles.
Inclusion criteria were as follows: 1) studies about the relationship between PTEN methylation and the clinicopathological significance of BC and 2) studies about the association of PTEN methylation and prognosis in patients with BC. The following were the exclusion criteria: 1) studies that investigated the association between PTEN protein expression and clinicopathological significance, 2) articles that failed to show the diagnosis of histologic categories of BC, 3) studies that utilized the same population or overlapping database, and 4) studies that utilized cell lines or mice. Eight articles were eligible for inclusion in this meta-analysis. This meta-analysis was conducted in accordance with Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.
Data extraction and study assessment
Two authors evaluated the included studies independently. Any disagreement was discussed and resolved by consensus. A standardized form was used to extract the data. The following information was collected from each study: first author’s name, year of publication, geographical location, sample size of histologic categories of BC, ER status, stage of BC, HER-2 status, and detective method of PTEN methylation.
Statistics analysis
Odds ratios (ORs) with their 95% confidence intervals (CIs) were calculated. Heterogeneity among studies was evaluated using the Cochran’s Q-statistic and I2 tests. A fixed-effects model was used for I2<50%, while a random-effects model was used for I2>50%. Since all studies in the analysis were not equally precise, some studies were more precise than others we assigned more weight to the studies that contained more clinical information or data. In the fixed-effects analysis, each study was weighted by the inverse of its variance. In the random-effects analysis also, each study was weighted by the inverse of its variance. The difference is that the variance in random model included the original (within-studies) variance plus the between-studies variance, tau-squared. The analysis was conducted to compare the frequency of PTEN promoter hypermethylation between DCIS and normal tissue, IDC and normal tissue, and IDC and DCIS. In addition, we evaluated the correlation between PTEN promoter hypermethylation and ER, HER-2 status, and frequency of PTEN promoter hypermethyaltion in different stages. Two-sided statistical tests and P-value were used. Publication bias was evaluated using visual inspection funnel plots. All analysis was conducted with Review Manager 5.2.
Results
The meta-analysis included eight studies and a total of 923 patients (Figure 1). The basic characteristics are summarized in Table 1.
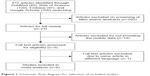  | Figure 1 Schematic flow diagram for selection of included studies. |
The frequency of PTEN promoter hypermethylation was significantly higher in DCIS than in normal breast tissues and the pooled OR was 22.53 (95% CI 4.35–116.72, z=3.71, P=0.0002, I2=50%; Figure 2). PTEN promoter hypermethylation was significantly correlated to the risk of IDC (OR =22.86, 95% CI 11.09–47.09, z=8.48, P<0.00001, I2=45%; Figure 3). The frequency of PTEN promoter hypermethylation was similar between IDC and DCIS (OR =1.39, 95% CI 0.50–3.87, z=0.63, P=0.53, I2=64%; Figure 4). In addition, we found that the frequency of PTEN promoter hypermethylation was not significantly associated with stages of BC (OR =1.80, 95% CI 0.44–7.37, z=0.82, P=0.41, I2=92%; Figure 5). The rate of PTEN promoter hypermethylation was similar between ER-positive and ER-negative status in patients with BC (OR =1.20, 95% CI 0.82–1.77, z=0.93, P=0.35, I2=32%; Figure 6). No significant difference in the frequency of PTEN promoter hypermethylation was observed between HER-2-positive and HER-2-negative BC (OR =1.27, 95% CI 0.65–2.49, z=0.69, P=0.49, I2=53%; Figure 7).
  | Figure 2 Forest plot for PTEN promoter hypermethylation in DCIS and normal breast tissue. |
  | Figure 3 Forest plot for PTEN promoter hypermethylation in IDC and normal breast tissue. |
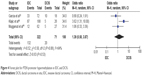  | Figure 4 Forest plot for PTEN promoter hypermethylation in IDC and DCIS. |
  | Figure 5 Forest plot for PTEN promoter hypermethylation in different stages of BC. |
  | Figure 6 Forest plot for PTEN promoter hypermethylation in ER-positive and ER-negative BC. |
The Newcastle Ottawa Quality Assessment Scale (NOQAS) was used to assess the methodological quality of the included studies. This scale was used to allocate a total of nine points for the quality of selection (four points), comparability (two points), and exposure (three points). The NOQAS scores ranged from 0 to 9, and a score ≥7 indicates a good quality. Three studies scored eight points and five scored seven points (Table 2). A sensitivity analysis was performed by omitting a study at a time. The results were not significantly changed after removing a study at a time, indicating the stability of present analyses. The funnel charts were largely symmetric (Figure 8), suggesting no publication biases existed in the meta-analysis of PTEN promoter hypermethylation and clinicopathological features.
Discussion
BC is the most common form of malignant disease in women worldwide and is also the principal cause of cancer-related mortality globally. It is particularly important to identify molecular biomarkers for the early detection of the disease and development of appropriate treatment strategies.
PTEN is a dual lipid/protein phosphatase that regulates cell cycle progression, survival, cell growth, angiogenesis, and genomic stability through PI3K/Akt signaling pathway.22–24 Its expression is modulated by germline and somatic PTEN mutation, genomic deletion, and promoter methylation silencing in many primary and metastatic tumors.18,25–28 Germline mutations throughout the PTEN coding region were found in PTEN hamartoma tumor syndrome (PHTS), which is a rare disease, including the previously named Cowden syndrome and Bannayan-Rilev-Ruvalcaba syndrome.29 PHTS patients have a high lifetime risk of developing breast, thyroid, or endometrial cancer due to PTEN silencing.30 Lifetime risks of developing breast, ovary, colorectal, and kidney cancers and melanoma increase in patients with germline PTEN mutations.31 PTEN promoter hypermethylation was reported in cervical cancer,32 colorectal cancer,33,34 esophageal squamous cell carcinoma,35 gastric cancer,36,37 hepatocellular carcinomas,38 head and neck cancer,39 lung cancer,40–42 and ovarian cancer.43 A frequency of less than 4% PTEN intragenic mutations were observed in sporadic BC.44–47 Therefore, PTEN promoter hypermethylation has been considered to be one of the most important mechanisms of inactivation. Several studies have reported more frequent PTEN promoter hypermethylation in DCIS than in normal breast tissue with inconsistent rate.20,48,49 We pooled those studies together and more precisely evaluated the frequency of PTEN promoter hypermethylation in DCIS and normal breast tissue. Our results showed PTEN promoter hypermethylation was significantly correlated with the risk of DCIS, suggesting PTEN promoter hypermethylation is an early event during breast carcinogenesis. Barekati et al investigated the promoter methylation of PTEN in triple-matched samples from BC, including cancerous tissue, matched adjacent normal tissue and serum samples, and found that the levels of PTEN methylation in tumor tissue and serum of patients was significantly increased compared to those in the normal breast tissue and serum, respectively.50 These data indicate that PTEN hypermethylation is a valuable biomarker for diagnosis of BC.
Interestingly, PTEN loss is more frequently observed in triple-negative BC than other subtypes of BC.51,52 Lately, Beg et al have reported that loss of PTEN expression correlated with invasive behavior and poor prognosis in Middle Eastern triple-negative BC, a type of high-grade BC with poor response to treatment.53 Thus, PTEN could be a novel target for the development of effective therapy, especially for patients with triple-negative BC. Further investigations need to be carried out in future. Additionally, the frequency of PTEN promoter hypermethylation in IDC was significantly increased compared to normal breast tissue. However, there was no significant difference between DIC and DCIS, and moreover PTEN was not frequently hypermethylated in early stage than late stage of BC patients, suggesting PTEN does not exhibit stage-dependent methylation pattern. Additional studies with a large number of patients are needed to clarify the relationship between PTEN promoter hypermethylation rate and tumor stages in the future.
In this study, we evaluated PTEN promoter hypermethylation in ER-positive versus ER-negative BC and showed similar frequency of hypermethylation, which is in line with previous studies.20,54 Additionally, HER-2, a transmembrane tyrosine kinase receptor, belongs to a family of epidermal growth factor receptors structurally related to epidermal growth factor receptor.55 HER-2 overexpression is observed in 20%–30% of BC cases and has been recognized as a poor marker of prognosis.56,57 Klajic et al found significantly higher z-scores of PTEN promoter hypermethylation in HER-2-positive than in HER-2-negative BCs.48 In the present study, there was no significant difference in the rate of PTEN promoter hypermethylation between HER-2-positive and HER-2-negative BCs. Due to the limited power of present study, more investigations need to be performed in the future. The present study included only the articles published in English or Chinese, and excluded relevant studies published in other languages. Therefore, our results should be interpreted with caution when applied to the general populations.
In summary, PTEN promoter is more frequently hypermethylated in DCIS and IDC compared to normal breast tissue, suggesting that PTEN is a valuable biomarker for diagnosis of BC. The frequency of PTEN promoter hypermethylation is similar between IDC and DCIS, indicating PTEN does not exhibit stage-dependent methylation pattern. In addition, it is likely that PTEN promoter hypermethylation is not significantly correlated with ER or HER-2 status in patients with BC.
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2013;63:11–30. | ||
Polyak K. Breast cancer: origins and evolution. J Clin Invest. 2007;117:3155–3163. | ||
Bombonati A, Sgroi DC. The molecular pathology of breast cancer progression. J Pathol. 2011;223:307–317. | ||
Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. | ||
Esteller M. Cancer Epigenetics for the 21st Century: What’s Next? Genes Cancer. 2011;2:604–606. | ||
Atalay C. Epigenetics in breast cancer. Exp Oncol. 2013;35:246–249. | ||
Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. | ||
Rasheed BK, Stenzel TT, McLendon RE, et al. PTEN gene mutations are seen in high-grade but not in low-grade gliomas. Cancer Res. 1997;57:4187–4190. | ||
Mikhail M, Velazquez E, Shapiro R, et al. PTEN expression in melanoma: relationship with patient survival, Bcl-2 expression, and proliferation. Clin Cancer Res. 2005;11:5153–5157. | ||
Yao YJ, Ping XL, Zhang H, et al. PTEN/MMAC1 mutations in hepatocellular carcinomas. Oncogene. 1999;18:3181–3185. | ||
Bruni P, Boccia A, Baldassarre G, et al. PTEN expression is reduced in a subset of sporadic thyroid carcinomas: evidence that PTEN-growth suppressing activity in thyroid cancer cells mediated by p27kip1. Oncogene. 2000;19:3146–3155. | ||
Tashiro H, Blazes MS, Wu R, et al. Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res. 1997;57:3935–3940. | ||
Sakai A, Thieblemont C, Wellmann A, Jaffe ES, Raffeld M. PTEN gene alterations in lymphoid neoplasms. Blood. 1998;92:3410–3415. | ||
Bose S, Crane A, Hibshoosh H, Mansukhani M, Sandweis L, Parsons R. Reduced expression of PTEN correlates with breast cancer progression. Hum Pathol. 2002;33:405–409. | ||
Perren A, Weng LP, Boag AH, et al. Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast. Am J Pathol. 1999;155:1253–1260. | ||
Tsutsui S, Inoue H, Yasuda K, et al. Reduced expression of PTEN protein and its prognostic implications in invasive ductal carcinoma of the breast. Oncology. 2005;68:398–404. | ||
Shetty PJ, Pasupuleti N, Chava S, Nasaruddin K, Hasan Q. Altered transcription and expression of PTEN in breast tumors: is it regulated by hypermethylation? Breast Dis. 2011;33:27–33. | ||
Jones N, Bonnet F, Sfar S, et al. Comprehensive analysis of PTEN status in breast carcinomas. Int J Cancer. 2013;133:323–334. | ||
Tserga A, Michalopoulos NV, Levidou G, et al. Association of aberrant DNA methylation with clinicopathological features in breast cancer. Oncol Rep. 2102;27:1630–1638. | ||
Garcia JM, Silva J, Pena C, et al. Promoter methylation of the PTEN gene is a common molecular change in breast cancer. Genes Chromosomes Cancer. 2001;41:117–124. | ||
Khan S, Kumagai T, Vora J, et al. PTEN promoter is methylated in a proportion of invasive breast cancers. Int J Cancer. 2004;112:407–410. | ||
Li L, Ross AH. Why is PTEN an important tumor suppressor? J Cell Biochem. 2007;102:1368–1374. | ||
Stambolic V, MacPherson D, Sas D, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. | ||
Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11:289–301. | ||
Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2001;2:489–501. | ||
Parsons DW, Wang TL, Samuels Y, et al. Colorectal cancer: mutations in a signalling pathway. Nature. 2005;436:792. | ||
Kechagioglou P, Papi RM, Provatopoulou X, et al. Tumor suppressor PTEN in breast cancer: heterozygosity, mutations and protein expression. Anticancer Res. 2014;34:1387–1400. | ||
Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. | ||
Orloff MS, Eng C. Genetic and phenotypic heterogeneity in the PTEN hamartoma tumour syndrome. Oncogene. 2008;27:5387–5397. | ||
Blumenthal GM, Dennis PA. PTEN hamartoma tumor syndromes. Eur J Hum Genet. 2008;16:1289–1300. | ||
Tan MH, Mester JL, Ngeow J, et al. Lifetime cancer risks in individuals with germline PTEN mutations. Clin Cancer Res. 2012;18:400–407. | ||
Qi Q, Ling Y, Zhu M, et al. Promoter region methylation and loss of protein expression of PTEN and significance in cervical cancer. Biomed Rep. 2014;2:653–658. | ||
Goel A, Arnold CN, Niedzwiecki D, et al. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res. 2004;64:3014–3021. | ||
Lin PC, Lin JK, Lin HH, et al. A comprehensive analysis of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) loss in colorectal cancer. World J Surg Oncol. 2015;13:186. | ||
Pan QF, Li WT, Dong HC, et al. PTEN hypermethylation profiles of Chinese Kazakh patients with esophageal squamous cell carcinoma. Dis Esophagus. 2014;27:396–402. | ||
Kang YH, Lee HS, KimWH. Promoter methylation and silencing of PTEN in gastric carcinoma. Lab Invest. 2002;82:285–291. | ||
Kang GH, Lee S, Kim WH, et al. Epstein-barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am J Pathol. 2002;160:787–794. | ||
Wang L, Wang WL, Zhang Y, Guo SP, Zhang J, Li QL. Epigenetic and genetic alterations of PTEN in hepatocellular carcinoma. Hepatol Res. 2007;37:389–396. | ||
Li J, Gong P, Lyu X, Yao K, Li X, Peng H. Aberrant CpG island methylation of PTEN is an early event in nasopharyngeal carcinoma and a potential diagnostic biomarker. Oncol Rep. 2014;31:2206–2212. | ||
Soria JC, et al. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res. 2001;8:1178–1184. | ||
Marsit CJ, Lee HY, Lee JI, et al. PTEN expression in non-small-cell lung cancer: evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum Pathol. 2005;36:768–776. | ||
Buckingham L, Penfield Faber L, Kim A, et al. PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer. 2010;126:1630–1639. | ||
Yang HJ, Liu VW, Wang Y, Tsang PC, Ngan HY. Differential DNA methylation profiles in gynecological cancers and correlation with clinico-pathological data. BMC Cancer. 2006;6:212. | ||
Rhei E, Kang L, Bogomolniy F, Federici MG, Borgen PI, Boyd J. Mutation analysis of the putative tumor suppressor gene PTEN/MMAC1 in primary breast carcinomas. Cancer Res. 1997;57:3657–3659. | ||
Bose S, Wang SI, Terry MB, Hibshoosh H, Parsons R. Allelic loss of chromosome 10q23 is associated with tumor progression in breast carcinomas. Oncogene. 1998;17:123–127. | ||
Feilotter HE, Coulon V, McVeigh JL, et al. Analysis of the 10q23 chromosomal region and the PTEN gene in human sporadic breast carcinoma. Br J Cancer. 1999;79:718–723. | ||
Millis SZ, Gatalica Z, Winkler J, et al. Predictive biomarker profiling of >6000 breast cancer patients shows heterogeneity in TNBC, with treatment implications. Clin Breast Cancer. 2015;15:473–481. | ||
Klajic J, Fleischer T, Dejeux E, et al. Quantitative DNA methylation analyses reveal stage dependent DNA methylation and association to clinico-pathological factors in breast tumors. BMC Cancer. 2013;13:456. | ||
Muggerud AA, Rønneberg JA, Wärnberg F, et al. Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res. 2010;12:R3. | ||
Barekati Z, Radpour R, Kohler C, et al. Methylation profile of TP53 regulatory pathway and mtDNA alterations in breast cancer patients lacking TP53 mutations. Hum Mol Genet. 2010;19:2936–2946. | ||
Dean SJ, Perks CM, Holly JM, et al. Loss of PTEN expression is associated with IGFBP2 expression, younger age, and late stage in triple-negative breast cancer. Am J Clin Pathol. 2014;141:323–333. | ||
Song CH, Park SY, Eom KY, et al. Potential prognostic value of heat-shock protein 90 in the presence of phosphatidylinositol-3-kinase overexpression or loss of PTEN, in invasive breast cancers. Breast Cancer Res. 2010;12:R20. | ||
Beg S, Siraj AK, Prabhakaran S, et al. Loss of PTEN expression is associated with aggressive behavior and poor prognosis in Middle Eastern triple-negative breast cancer. Breast Cancer Res Treat. 2015;151:541–553. | ||
Zhang HY, Liang F, Jia ZL, Song ST, Jiang ZF. Mutation, methylation and expression in breast cancer patients. Oncol Lett. 2013;6:161–168. | ||
Yamamoto T, Ikawa S, Akiyama T, et al. Similarity of protein encoded by the human c-erb-B-2 gene to epidermal growth factor receptor. Nature. 1986;319:230–234. | ||
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. | ||
Barnes DM. c-erbB-2 amplification in mammary carcinoma. J Cell Biochem Suppl. 1993;17G:132–138. | ||
Zhao YF, Shen SP, Jiang JY, Geng H, Guo JG, Xie LP. [Methylation and expression of gene p16INK4a and RB in breast carcinom]. Zhonghua Bing Li Xue Za Zhi. 2010;39(6):377–381. Chinese. | ||
Yari K, Payandeh M, Rahimi Z. Association of the hypermethylation status of PTEN tumor suppressor gene with the risk of breast cancer among Kurdish population from Western Iran. Tumour Biol. 2016;37(6):8145–8152. | ||
Siddiqui S, Akhter N, Deo SV, Shukla NK, Husain SA. A study on promoter methylation of PTEN in sporadic breast cancer patients from North India. Breast Cancer. Epub 2016 Jan 11. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




