Back to Journals » OncoTargets and Therapy » Volume 8
The association between the migration inhibitory factor -173G/C polymorphism and cancer risk: a meta-analysis
Authors Zhang X, Weng W, Xu W, Wang Y, Yu W, Tang X, Ma L, Pan Q, Wang J, Sun F
Received 17 August 2014
Accepted for publication 14 January 2015
Published 10 March 2015 Volume 2015:8 Pages 601—613
DOI https://doi.org/10.2147/OTT.S72795
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Faris Farassati
Xiao Zhang,1 Wenhao Weng,1 Wen Xu,2 Yulan Wang,1 Wenjun Yu,1 Xun Tang,1 Lifang Ma,1 Qiuhui Pan,3 Jiayi Wang,1 Fenyong Sun1
1Department of Clinical laboratory medicine, Shanghai Tenth People’s Hospital of Tongji University, 2Department of Clinical laboratory medicine, Zhongshan Hospital, Fudan University, 3Department of Central Laboratory, Shanghai Tenth People’s Hospital of Tongji University, Shanghai, People’s Republic of China
Abstract: Previous studies have suggested that macrophage migration inhibitory factor (MIF) -173G/C polymorphism may be associated with cancer risk. However, previous research has demonstrated conflicting results. Therefore, we followed the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines and the meta-analysis on genetic association studies checklist, and performed a meta-analysis to investigate the association between MIF -173G/C polymorphisms and the risk of cancer. Odds ratios (ORs) and corresponding 95% confidence intervals (CIs) were combined to measure the association between MIF promoter polymorphisms and cancer risk. The pooled ORs were performed for the dominant model, recessive model, allelic model, homozygote comparison, and heterozygote comparison. The publication bias was examined by Begg’s funnel plots and Egger’s test. A total of ten studies enrolling 2,203 cases and 2,805 controls met the inclusion criteria. MIF (-173G/C) polymorphism was significantly associated with increased cancer risk under the dominant model (OR=1.32, 95%, CI=1.00–1.74, P=0.01) and the heterozygote comparison (OR=1.38, CI=1.01–1.87, P=0.04). In subgroup analysis, MIF polymorphism and prostate were related to increased risk of prostate and non-solid cancer. In conclusion, MIF polymorphism was significantly associated with cancer risk in heterozygote comparison. The MIF -173G/C polymorphism may be associated with increased cancer risk.
Keywords: MIF, SNP, systematic review, cancer susceptibility
Introduction
Macrophage migration inhibitory factor (MIF) was first identified nearly 50 years ago and has been used as a cytokine and an enzyme.1,2 MIF is a member of the transferring growth factor-β (TGF-β) super family, which is expressed by a broad variety of cells, including B- and T-lymphocytes as well as endocrine, endothelial, and epithelial cells of diverse histogenetic origin.3 Presently, MIF is considered to play an important role in the pro- and anti-inflammatory response to infection since it is constitutively expressed and acts as an upstream regulator of many other inflammatory cytokines.4,5
Recently, several studies have shown that MIF can promote tumor growth and viability by modulating immune responses and supporting tumor-associated angiogenesis.6 A few experiments suggested that MIF mRNA and MIF protein are overexpressed in a number of cancers.7 Tan et al reported that MIF is upregulated in patients with pancreatic cancer and causes dysfunction of insulin secretion in β-cells.8 Krockenberger et al reported that MIF is clearly overexpressed on the protein level in invasive cervical cancer compared to cervical dysplasia.9 Two polymorphisms in the promoter region of MIF have been reported in the past. One is a single nucleotide polymorphism (SNP) at the nucleotide position −173 (G to C)10 and the other is a tetranucleotide CATT repeat beginning at position −794.11 The association between these two polymorphisms and diseases has been extended to several inflammatory conditions including Graves’ disease,12 idiopathic thrombocytopenic purpura,13 and Vogt-Koyanagi-Harada (VKH) syndrome.14 These studies indicate that these two polymorphisms of MIF are associated with inflammatory diseases. Similarly, some studies have reported that the polymorphism of MIF resulted in an increased risk of cancer. With new studies about the polymorphism of MIF and the risk of cancer emerging, there has been no meta-analysis conducted regarding the association between MIF promoter polymorphism and the risk of cancer in recent times. The aim of this study is to perform a meta-analysis of all available studies that analyze the association between the polymorphism of MIF promoter and the risk of cancer.
Materials and methods
Literature search
The preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement (Figure S1) and the meta-analysis on genetic association studies checklist (Figure S2) were followed in our meta-analysis. A comprehensive search of EMBASE, PubMed, Web of Science, OVID, Cochrane Library, and China National Knowledge Infrastructure (CNKI) was done from database inception to July 22, 2014 without language restriction. The search strategy was “macrophage migration inhibitory factor or MIF” and “polymorphism or variant or mutation or genotype.” To complete our research, we also studied the review articles and references of retrieved articles manually. The literature review was performed independently by X Zhang and J Wang and the disagreements were resolved through consensus by all the authors.15,16
Selection criteria
Studies were included in the meta-analysis if the following inclusion criteria were satisfied: 1) case-control studies focused on association between the MIF promoter polymorphism and cancer risk, 2) studies enrolled more than 30 patients, 3) studies provided sufficient data to estimate the odds ratio (OR) and 95% confidence intervals (CIs) according to MIF promoter polymorphism, and 4) when study patients overlapped with patients in other included studies, we selected the first study published. The two researchers (J Wang and X Zhang) independently read the titles and abstracts and excluded the uncorrelated studies; then the full-texts were examined by our review team. The studies were selected according to the inclusion criteria.15,16
Data abstraction
Two independent reviewers (X Zhang and J Wang) extracted the following information: authors, year of publication, country, tumor type, number of cases and controls analyzed, mean value of age, source of controls (hospital-based controls or population-based controls), and genotyping method. If both univariate and multivariate analyses were reported, we utilized the multivariate analysis because it involves observation and analysis of more than one statistical outcome variable at a time thus is more accurate. If articles provided insufficient data (missing data, inconsistencies, or any other uncertainties), we attempted to contact the first and corresponding authors for necessary information via telephone or email.15,16
Statistical analysis
ORs and corresponding 95% CIs were combined to measure the association between MIF promoter polymorphisms and cancer risk. Hardy–Weinberg equilibrium (HWE) for each study was determined by the chi-square test. The pooled ORs were calculated for the allelic model (mutation [M] allele versus [vs] wild [W] allele), dominant model (WM + MM vs WW), recessive model (MM vs WM + WW), homozygote comparison (MM vs WW), and heterozygote comparison (WM vs WW) respectively, and P<0.05 denoted statistical significance. Statistical heterogeneity among the studies was evaluated using the Q-test and I2-test. When heterogeneity among the studies was observed, the pooled OR was calculated by random-effect models. Sensitivity analyses were performed to identify the potential influence of the individual data set to the pooled ORs. Subgroup analyses were conducted with respect to cancer type and source of controls. The statistical significance was analyzed by Student’s t-test. These analyses were performed by Review Manager Version 5.1 software (http://ims.cochrane.org/revman). Both Begg’s and Egger’s tests was performed using R (http://cran.r-project.org/bin/windows/base).15,16
Results
Characteristics of identified studies
Following an initial search, 166 studies were retrieved from PubMed; 233 studies from EMBASE; 313 studies from OVID; 266 studies from Web of Science; 50 studies from Cochrane Library; 532 studies from CNKI; and five additional review articles were added to make our search comprehensive. After duplicated records were removed, 878 published studies were identified. We excluded 780 unrelated studies by reading the titles and abstracts. Next, we downloaded the full-text of the remaining 98 studies and excluded 65 unrelated studies. Of the remaining 33 studies considered for performing the meta-analysis, some studies were found to report incomplete data or report other associations between MIF and cancer. We tried our best to communicate with the first and corresponding authors to get the necessary data. Some authors were able to provide the necessary data for our study, while others did not. Ultimately, after further reviewing in detail, ten studies were included in our meta-analysis.17–26 Figure 1 shows in detail the selection process. These ten studies were published between 2005 and 2014. There were 2,203 cases and 2,805 controls included in our meta-analysis. Studies were carried out in People’s Republic of China, Taiwan, Japan, Iran, Italy, and USA. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) was used in seven studies.17,18,20,21,23,25,26 One study used polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP).24 The other two studies employed denaturing high-performance liquid chromatography (DHLPC) wave analysis19 and a Genetic Analyzer,22 respectively. Three studies assessed prostate cancer;20,22,26 three studies assessed leukemia17,19,25 and one each for gastric cancer,24 cervical cancer,18 colorectal cancer,21 and bladder cancer.23 The genotype distribution in one study deviated from HWE.26 The main characteristics of all the included studies are listed in Table 1.
  | Figure 1 Flow diagram summarizing the selection of eligible studies. |
Meta-analysis
Overall, ten prospective studies enrolling 2,203 cases and 2,805 controls were included in our meta-analysis. A statistically significant association between MIF (−173G/C) polymorphism and cancer risk was found under the dominant model (OR=1.32, CI=1.00–1.74, P=0.01) (Figure 2) and the heterozygote comparison (OR=1.38, CI=1.01–1.87, P=0.04) (Figure S3). There was no statistical significant association under the recessive model (OR=0.98, 95% CI 0.67–1.45, P=0.93) (Figure S4), homozygote comparison (OR=1.02, 95% CI 0.64–1.63, P=0.93) (Figure S5), and allelic model (OR=1.32, 95% CI 1.00–1.74, P=0.05) (Figure S6). Furthermore, in our subgroup analysis, a significant association was found in the prostate group under the dominant model (OR=3.34, 95% CI 2.24–4.97, P<0.001), allelic model (OR=2.94, 95% CI 1.91–4.54, P<0.001), and heterozygote comparison (OR=2.39, 95% CI 1.65–3.47, P<0.001). MIF (−173G/C) polymorphism was also significantly associated with non-solid cancer risk under the dominant model (OR=1.27, 95% CI 1.03–1.56, P=0.03) and heterozygote comparison (OR=1.32, 95% CI 1.06–1.63, P=0.01). Table S1 presents the results of overall and subgroup analyses.
  | Figure 2 Forest plot of MIF –173G/C polymorphism and cancer risk in dominant model. |
Sensitivity analysis
We performed sensitivity analysis by omitting one study at a time and calculating the pooled ORs again. However, the results did not show any significant statistical differences when studies were omitted. Therefore, the stability of the study was not influenced by any individual study. Table S2 presents the sensitivity analysis in the dominant model.
Publication bias
Both Begg’s funnel plot and Egger’s test were carried out to evaluate the publication bias of the studies. The results are presented in Figure 3 and Table 2. Publication bias was found under the dominant model (P=0.0286) according to Begg’s funnel plot. When Egger’s test was performed, publication bias was found under the recessive model (P=0.0075) and homozygote comparison (P=0.03). Results indicate that there may be publication bias existing in our meta-analysis. Table 2 presents the results of Begg’s funnel plot and Egger’s test under the five genetic models.
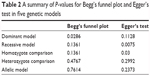  | Table 2 A summary of P-values for Begg’s funnel plot and Egger’s test in five genetic models |
Discussion
In our meta-analysis, ten studies enrolling 2,203 cases and 2,805 controls were included. The results indicated that MIF −173G/C polymorphism was significantly associated with cancer risk.
MIF is known as a major regulator of inflammation and a central upstream mediator of innate immune response, and functions as a key mediator to counter-regulate the inhibitory effects of glucocorticoids within the immune system.27 There are numerous studies suggesting that MIF polymorphism might be associated with the risk of immune disease. Liu et al reported that MIF polymorphism is associated with new-onset Graves’ disease in a Taiwanese Chinese population.12 Hao et al carried out a meta-analysis to investigate the association between MIF polymorphism and the risk of inflammatory bowel disease (IBD).28 They found that MIF −173G/C polymorphism contributed to the susceptibility of IBD.
MIF is also involved in cancer growth and progression. The elevated MIF and mRNA levels have been observed in many tumor cells and pre-tumor states. Krockenberger et al found that MIF was significantly overexpressed on both the protein level and the mRNA level in invasive cervical cancer and MIF protein was overexpressed in SiHA and CaSki cervical cancer cell lines.9 Huang et al reported that MIF expression levels in hepatocellular carcinoma tissues and cell lines were significantly up-regulated compared with adjacent normal tissues or a normal liver cell line.29 Moreover, several studies suggested that MIF polymorphism might be associated with the risk of cancer. Only one study reported that MIF −173G/C polymorphism is associated with a decreased risk of cancer.23 All the other studies reported the opposite conclusion. We also found a meta-analysis that investigated the association between the MIF −173G/C polymorphism and cancer risk.30 However, there were only five studies included in that meta-analysis, and the result was only under the dominant model. In recent times, some new studies have been emerging; for instance, Yuan et al reported that MIF −173G/C polymorphism is associated with decreased cancer risk.23 This conclusion contradicted with the conclusion in the previous meta-analysis. Therefore, we added new studies in our meta-analysis and calculated ORs in the dominant model, recessive model, homozygote comparison, heterozygote comparison, and allelic model. In our meta-analysis, we found that MIF −173G/C polymorphism is significantly associated with cancer risk in the dominant model (OR=1.32, 95% CI 1.00–1.74, P=0.01) and heterozygote comparison (OR=1.38, 95% CI 1.01–1.87, P=0.04). There were no significant associations between MIF −173G/C polymorphism and cancer risk in the recessive model (OR=0.98, 95% CI 0.67–1.45, P=0.93), homozygote comparison (OR=1.02, 95% CI 0.64–1.63, P=0.93), and allelic model (OR=1.32, 95% CI 1.00–1.74, P=0.05). Drawing from these results, we conclude from our meta-analysis that MIF −173G/C polymorphism might increase the risk of cancer.
There are several limitations in our meta-analysis. First, publication bias exists in the current meta-analysis. If the future studies find that MIF polymorphism was not associated with cancer risk, then publication bias might cause false outcomes. Second, there were some studies lacking in necessary data to calculate ORs under different genetic models. Although we had tried our best to communicate with the first and corresponding authors, some were unable to reply. Third, the patients included in the meta-analysis were limited. It was difficult for us to perform subgroup analyses and obtain specific results. Additionally, only papers published in English or Chinese were included in our meta-analysis, and eligible studies written in other languages that could have fulfilled our study criterion were not included.
Conclusion
Our meta-analysis concluded that MIF −173G/C polymorphism might increase the risk of cancer. Given the above limitations, more studies are needed to confirm the association between MIF polymorphism and the risk of cancer.
Acknowledgment
This study was supported by Natural Science Foundation of People’s Republic of China (Grant Nos: 81201363 and 81301689), and the Climbing training program (assigned to Jiayi Wang) from Shanghai Tenth People’s Hospital.
Disclosure
The authors report no conflicts of interest in this work.
References
Bloom BR, Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153(3731):80–82. | ||
Lolis E, Bucala R. Macrophage migration inhibitory factor. Expert Opin Ther Targets. 2003;7(2):153–164. | ||
He M, Metz C, Cheng KF, et al. Novel arylazoarylmethane as potential inhibitor of macrophage migration inhibitory factor. Arch Pharm. 2014;347(2):104–107. | ||
Grieb G, Merk M, Bernhagen J, Bucala R. Macrophage migration inhibitory factor (MIF): a promising biomarker. Drug News Perspect. 2010;23(4):257–264. | ||
Vujicic M, Senerovic L, Nikolic I, Saksida T, Stosic-Grujicic S, Stojanovic I. The critical role of macrophage migration inhibitory factor in insulin activity. Cytokine. 2014;69(1):39–46. | ||
Yasasever V, Camlica H, Duranyildiz D, Oguz H, Tas F, Dalay N. Macrophage migration inhibitory factor in cancer. Cancer Invest. 2007; 25(8):715–719. | ||
Armstrong ME, Gantier M, Li L, et al. Small interfering RNAs induce macrophage migration inhibitory factor production and proliferation in breast cancer cells via a double-stranded RNA-dependent protein kinase-dependent mechanism. J Immunol. 2008;180(11):7125–7133. | ||
Tan L, Ye X, Zhou Y, et al. Macrophage migration inhibitory factor is overexpressed in pancreatic cancer tissues and impairs insulin secretion function of beta-cell. J Transl Med. 2014;12:92. | ||
Krockenberger M, Engel JB, Kolb J, et al. Macrophage migration inhibitory factor expression in cervical cancer. J Cancer Res Clin Oncol. 2010;136(5):651–657. | ||
Amoli MM, Donn RP, Thomson W, et al. Macrophage migration inhibitory factor gene polymorphism is associated with sarcoidosis in biopsy proven erythema nodosum. J Rheumatol. 2002;29(8):1671–1673. | ||
Baugh JA, Chitnis S, Donnelly SC, et al. A functional promoter polymorphism in the macrophage migration inhibitory factor (MIF) gene associated with disease severity in rheumatoid arthritis. Genes Immun. 2002;3(3):170–176. | ||
Liu YH, Chen CC, Yang CM, Chen YJ, Tsai FJ. Dual effect of a polymorphism in the macrophage migration inhibitory factor gene is associated with new-onset Graves disease in a Taiwanese Chinese population. PLoS One. 2014;9(3):e92849. | ||
Lao W, Xiang Y, Fang M, Yang X. Polymorphism and expression of macrophage migration inhibitory factor does not contribute to glucocorticoid resistance in idiopathic thrombocytopenic purpura. Pharmazie. 2013;68(10):846–849. | ||
Zhang C, Liu S, Hou S, et al. MIF gene polymorphisms confer susceptibility to Vogt-Koyanagi-Harada syndrome in a Han Chinese population. Invest Ophthalmol Vis Sci. 2013;54(12):7734–7738. | ||
Zhang X, Weng W, Xu W, et al. Role of Bcl-2-938 C>A polymorphism in susceptibility and prognosis of cancer: a meta-analysis. Sci Rep. 2014; 4:7241. | ||
Zhang X, Weng W, Xu W, et al. Prognostic significance of interleukin 17 in cancer: a meta-analysis. Int J Clin Exp Med. 2014;7(10): 3258–3269. | ||
Ramireddy L, Lin CY, Liu SC, et al. Association study between macrophage migration inhibitory factor −173 polymorphism and acute myeloid leukemia in Taiwan. Cell Biochem Biophys. 2014;70(2): 1159–1165. | ||
Wu S, Lian J, Tao H, Shang H, Zhang L. Correlation of macrophage migration inhibitory factor gene polymorphism with the risk of early-stage cervical cancer and lymphatic metastasis. Oncol Lett. 2011;2(6): 1261–1267. | ||
Ziino O, D’Urbano LE, De Benedetti F, et al. The MIF −173G/C polymorphism does not contribute to prednisone poor response in vivo in childhood acute lymphoblastic leukemia. Leukemia. 2005;19(12): 2346–2347. | ||
Razzaghi M, Mazloomfard M, Lotfi B. Macrophage migration inhibitory factor gene −173*C polymorphism and risk of prostate cancer. Eur Urol Suppl. 2012;19(12):142. | ||
Ramireddy L, Chen WT, Peng CT, et al. Association between genetic polymorphism of the MIF gene and colorectal cancer in Taiwan. J Clin Lab Anal. 2014. | ||
Meyer-Siegler KL, Vera PL, Iczkowski KA, et al. Macrophage migration inhibitory factor (MIF) gene polymorphisms are associated with increased prostate cancer incidence. Genes Immun. 2007;8(8): 646–652. | ||
Yuan Q, Wang M, Wang M, Zhang Z, Zhang W. Macrophage migration inhibitory factor gene −173G>C polymorphism and risk of bladder cancer in southeast China: a case-control analysis. Mol Biol Rep. 2012;39(3):3109–3115. | ||
Arisawa T, Tahara T, Shibata T, et al. Functional promoter polymorphisms of the macrophage migration inhibitory factor gene in gastric carcinogenesis. Oncol Rep. 2008;19(1):223–228. | ||
Xue Y, Xu H, Rong L, et al. The MIF −173G/C polymorphism and risk of childhood acute lymphoblastic leukemia in a Chinese population. Leuk Res. 2010;34(10):1282–1286. | ||
Ding GX, Zhou SQ, Xu Z, et al. The association between MIF −173 G>C polymorphism and prostate cancer in southern Chinese. J Surg Oncol. 2009;100(2):106–110. | ||
Xu L, Li Y, Sun H, et al. Current developments of macrophage migration inhibitory factor (MIF) inhibitors. Drug Discov Today. 2013;18(11–12):592–600. | ||
Hao NB, He YF, Luo G, Yong X, Zhang Y, Yang SM. Macrophage migration inhibitory factor polymorphism and the risk of ulcerative colitis and Crohn’s disease in Asian and European populations: a meta-analysis. BMJ Open. 2013;3(12):e003729. | ||
Huang XH, Jian WH, Wu ZF, et al. Small interfering RNA (siRNA)-mediated knockdown of macrophage migration inhibitory factor (MIF) suppressed cyclin D1 expression and hepatocellular carcinoma cell proliferation. Oncotarget. 2014;5(14):5570–5580. | ||
Vera PL, Meyer-Siegler KL. Association between macrophage migration inhibitory factor promoter region polymorphism (−173 G/C) and cancer: a meta-analysis. BMC Res Notes. 2011;4:395. |
Supplementary materials
  | Table S2 The influence of individual study on ORs in dominant model |
  | Figure S1 Preferred reporting items for systematic reviews and meta-analyses (PRISMA) checklist |
  | Figure S2 Meta-analysis on genetic association studies checklist |
  | Figure S3 Forest plot of MIF –173G/C polymorphism and cancer risk in heterozygote comparison. |
  | Figure S4 Forest plot of MIF –173G/C polymorphism and cancer risk in recessive model. |
  | Figure S5 Forest plot of MIF –173G/C polymorphism and cancer risk in homozygote comparison. |
  | Figure S6 Forest plot of MIF –173G/C polymorphism and cancer risk in allelic model. |
References
Moher D, Liberati A, Tetzlaff J, Altman DG. The PRISMA Group (2009). Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 6(6):e1000097. | ||
Ramireddy L, Lin CY, Liu SC, et al. Association study between macrophage migration inhibitory factor-173 polymorphism and acute myeloid leukemia in Taiwan. Cell Biochem Biophys. 2014;70(2):1159–1165. | ||
Wu S, Lian J, Tao H, Shang H, Zhang L. Correlation of macrophage migration inhibitory factor gene polymorphism with the risk of early-stage cervical cancer and lymphatic metastasis. Oncol Lett. 2011;2(6):1261–1267. | ||
Ziino O, D'Urbano LE, De Benedetti F, et al. The MIF-173G/C polymorphism does not contribute to prednisone poor response in vivo in childhood acute lymphoblastic leukemia. Leukemia. 2005;19(12):2346–2347. | ||
Razzaghi M, Mazloomfard M, Lotfi B. Macrophage migration inhibitory factor gene-173*C polymorphism and risk of prostate cancer. Eur Urol Suppl. 2012;19(12):142. | ||
Ramireddy L, Chen WT, Peng CT, et al. Association between genetic polymorphism of the MIF gene and colorectal cancer in Taiwan. J Clin Lab Anal. 2014. | ||
Meyer-Siegler KL, Vera PL, Iczkowski KA, et al. Macrophage migration inhibitory factor (MIF) gene polymorphisms are associated with increased prostate cancer incidence. Genes Immun. 2007;8(8):646–652. | ||
Yuan Q, Wang M, Wang M, Zhang Z, Zhang W. Macrophage migration inhibitory factor gene -173G>C polymorphism and risk of bladder cancer in southeast China: a case-control analysis. Mol Biol Rep. 2012;39(3):3109–3115. | ||
Arisawa T, Tahara T, Shibata T, et al. Functional promoter polymorphisms of the macrophage migration inhibitory factor gene in gastric carcinogenesis. Oncol Rep. 2008;19(1):223–228. | ||
Xue Y, Xu H, Rong L, et al. The MIF-173G/C polymorphism and risk of childhood acute lymphoblastic leukemia in a Chinese population. Leuk Res. 2010;34(10):1282–1286. | ||
Ding GX, Zhou SQ, Xu Z, et al. The association between MIF-173 G>C polymorphism and prostate cancer in southern Chinese. J Surg Oncol. 2009;100(2):106–110. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



