Back to Journals » Cancer Management and Research » Volume 13
The Association Between PNPLA2 Methylation in Peripheral Blood and Early-Stage Lung Cancer in a Case–Control Study
Authors Qiao R, Li M, Zhong R , Wei Y, Wang J, Zhang Z, Wang L, Xu T, Wang Y, Dai L, Gu W , Han B, Yang R
Received 15 July 2021
Accepted for publication 20 September 2021
Published 15 October 2021 Volume 2021:13 Pages 7919—7927
DOI https://doi.org/10.2147/CMAR.S329629
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Kattesh Katti
Rong Qiao,1,* Mengxia Li,2,* Runbo Zhong,1 Yujie Wei,3 Jun Wang,3 Zheng Zhang,3 Ling Wang,3 Tian Xu,4 Yue Wang,1 Liping Dai,5 Wanjian Gu,4 Baohui Han,1 Rongxi Yang2,3
1Department of Pulmonary Medicine, Shanghai Chest Hospital, Shanghai Jiaotong University, Shanghai, 200030, People’s Republic of China; 2Department of Epidemiology and Biostatistics, School of Public Health, Nanjing Medical University, Nanjing, 210000, People’s Republic of China; 3Nanjing TANTICA Biotechnology Co. Ltd, Nanjing, 210000, People’s Republic of China; 4Department of Clinical Laboratory, Jiangsu Province Hospital of Chinese Medicine, Nanjing, 210000, People’s Republic of China; 5Henan Institute of Medical and Pharmaceutical Sciences, Academy of Medical Science, Zhengzhou University, Zhengzhou, 450052, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Rongxi Yang; Baohui Han Fax +86 25 86527613
Email [email protected]; [email protected]
Purpose: Lung cancer (LC) brings great burden to the society worldwide. Exploring novel biomarkers in vitro for the early detection of LC would be of great importance.
Patients and Methods: We measured DNA methylation levels of 21 CpG sites within Patatin-like phospholipase domain containing 2 (PNPLA2) gene in the peripheral blood of 168 early-stage LC cases (94.0% LC at stage I) and 187 age- and gender-matched cancer-free controls. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using logistic regression adjusted for covariates. Non-parametric tests were applied for the comparisons of stratified groups.
Results: Hypomethylation of PNPLA2_CpG_8,10 and hypermethylation of PNPLA2_CpG_9 were correlated to the early-stage LC with the ORs of 1.44 (95% CI: 1.06– 1.96, P = 0.018) and 0.82 (95% CI: 0.69– 0.98, P = 0.029), respectively. The associations were still significant for the very early-stage LC patients (stage I). Further gender- and age-stratified analyses indicated that the association between hypomethylation of PNPLA2_CpG_8,10 and LC existed only in females and in subjects younger than 55 years. In addition, the association between LC and hypermethylation of PNPLA2_CpG_6 and PNPLA2_CpG_9 was also observed in the younger population.
Conclusion: Taken together, our study has proved the hypothesis that the altered methylation in the peripheral blood may be correlated with the burden of cancer at an early stage. Here, we find a novel association between blood-based aberrant PNPLA2 methylation and LC at a very early stage and particularly for women at a younger age.
Keywords: lung cancer, DNA methylation, peripheral blood, biomarker
Introduction
It has been estimated that lung cancer (LC) accounted for more than 2.09 million new cases and 1.76 million deaths worldwide in 2018, making it the leading cause of cancer-related incidence and mortality in men and women.1 Data from the Global Burden of Disease study demonstrated that, from 1990 to 2017, the age-standardized death rates for LC showed a decreasing trend in men and an increasing trend in women worldwide, while raised in both men and women in China, indicating that LC in China represents a tremendous burden on the global epidemic.2 For non-small cell lung cancer (NSCLC) patients, a decline in the 5-year survival rates with the clinical stages was well documented, ranging from 92% at the stage IA1 to 6% at the stage IV.3
Therefore, other than the development of treatment and therapy for LC, early detection may be an alternative option for the easing of LC burden in China. A large randomized controlled trial showed an inspiring 20.0% decrease in mortality of persons at high risk for LC by low-dose computed tomographic (LDCT) screening, while the simultaneous high false-positive rate should also be concerned.4 Besides, overdiagnosis, cost-effectiveness, radiation risks, and abnormal opacities caused by other diseases were also uncertainties to be considered before conducting a screening with LDCT.5
Adjunctive methods to improve the precision on the basis of LDCT are urgently needed. Epigenetics, characterized by regulating gene expression without changes in the DNA sequence, performs a critical function in initiation and progression of many diseases, thus attracting accumulative interests in recent decades.6 DNA methylation is the most studied epigenetic modification. Altered DNA methylation in peripheral blood has been disclosed as a potential biomarker of LC, due to its superiority in biological stability and noninvasive sample collection.7 For example, hypomethylation of AHRR and F2RL3 gene in peripheral blood showed enhanced predictive value for LC risk in the prospective studies.8 Our previous case-control study also indicated that peripheral blood-based RAPSN hypomethylation was associated with early-stage LC.9
Patatin-like phospholipase domain containing 2 (PNPLA2) gene, also known as ATGL, encodes an enzyme catalyzing the rate-limiting step in the hydrolysis of triglycerides.10 PNPLA2 is related to many diseases in human, including neutral lipid storage disease with myopathy, breast cancer, hepatocellular cancer, and pancreatic ductal adenocarcinoma.11 PNPLA2 may be involved in the process of cancers via p-AKT signaling pathway, AMP-kinase and mTOR signaling pathway and other lipolysis-related events.11,12 Notably, a study reported that maternal prenatal folic acid supplementation may improve lipid metabolism in adult rats by elevated DNA methylation levels within ATGL and LPL promoter/first exon region.13 However, there is no report about the association between blood-based PNPLA2 methylation and cancers. In our present study, we aimed to investigate the association between altered PNPLA2 methylation in the peripheral blood and early-stage LC in a case-control study in the Chinese population.
Patients and Methods
Study Design and Population
This was a case-control study design. A total of 168 LC cases, determined by thoracic surgery and pathology, were collected from Shanghai Chest Hospital in 2018–2019. 187 age- and gender-matched healthy controls were consecutively collected from a physical examination center at Jiangsu Province Hospital of Chinese Medicine during the year of 2018–2019. No further inclusion criteria were applied for the controls. The median (interquartile range, IQR) age of LC cases and healthy controls were 57.5 (53.3-63.8) and 55.0 (51.0–63.0) years old, respectively. All the LC patients in the present study were diagnosed at very early stage (158 at stage I and 10 at stage II), with 64.3% males and 35.7% females. Additional clinical information of the LC patients, such as the tumor size, the tumor length and the involved lymph nodes, was described in detail in Table 1.
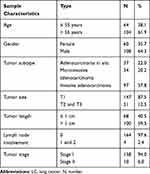 |
Table 1 Clinical Characteristics of LC Cases |
This study was conducted in accordance with the Declaration of Helsinki. The present study was approved by the Ethics Committee of the Shanghai Chest Hospital and Jiangsu Province Hospital of Chinese Medicine in China. Written informed consents were obtained from all the participants.
Sample Collection and Processing
Ethylenediamine tetraacetic acid (EDTA) tubes were used to collect peripheral blood. For LC patients, blood samples were taken before surgery and before any LC-related treatments. All the blood samples were kept at 4°C for up to 24 hours after the blood extraction and stored at −80°C for future usage. DNA Extraction Kit (TANTICA, Nanjing, China) was used for the extraction of DNA from blood. All the cases and controls were processed in parallel.
Bisulfite Conversion
DNA of each sample was converted by a bisulfite reaction using EZ-96 DNA Methylation Gold Kit (Zymo Research, Orange, USA) according to the manufacturer’s specifications. The bisulfite treatment converts non-methylated cytosine (C) at the CpG site to uracil (U), while methylated cytosine remains intact.
Matrix-Assisted Laser Desorption Ionization Time-of-Flight (MALDI-TOF) Mass Spectrometry
Polymerase chain reaction (PCR) was used to amplify a 355 bp amplicon in PNPLA2 gene covering 21 CpG sites. There are no single nucleotide polymorphisms (SNPs) located at the primer regions or overlapped with any of these CpG sites. The designed bisulfite-specific primers for PNPLA2 gene are available upon request. The methylation levels of PNPLA2 CpG sites were determined by MALDI-TOF mass spectrometry (Agena Bioscience, San Diego, California, US) in a quantitative manner as described before14 and as presented in Supplementary Figure 1. In brief, the PCR products were incubated with Shrimp Alkaline Phosphatase (SAP) and treated by the T cleavage assay (Agena Bioscience, San Diego, California, U.S.), and further cleaned by Resin. The final products were transferred to a SpectroCHIP G384 and detected by the MassARRAY spectrometry (Agena Bioscience, San Diego, California, U.S.). The quantitative methylation level of each CpG site was collected by SpectroACQUIRE v3.3.1.3 software and visualized by EpiTyper v1.3 software. A total of 21 CpG sites were detected in the amplicon of PNPLA2, of which 20 CpG sites were measurable, and the PNPLA2_CpG_19 CpG site was excluded own to low quality of signal. These 20 measurable CpG loci yielded 13 signal peaks by the mass spectrometry, whereas most of the signal peaks contained only one CpG site. When two or more CpG sites were located at the same fragment, the methylation intensity was presented as the average intensity of these CpG sites, such as PNPLA2_CpG_11,12, PNPLA2_CpG_14,15,16,17 and PNPLA2_CpG_20,21. When two CpG sites shared the same mass, we showed their methylation intensities as combined briefly, for example, PNPLA2_CpG_2,3 stands for PNPLA2_CpG_2 and PNPLA2_CpG_3, and PNPLA2_CpG_8,10 stands for PNPLA2_CpG_8 and PNPLA2_CpG_10. The hypo- and hypermethylation was defined by the methylation levels of the cases compared to the controls. When the methylation levels of cases were lower than the controls, hypomethylation was referred. When the methylation levels of cases were higher than the controls, hypermethylation was referred.
Statistical Analyses
All statistical analyses were carried out using IBM SPSS Statistics 25.0 version (Chicago, IL, USA). Binary logistic regression was performed with the adjustment of age, gender and experimental batches. Odds ratios (ORs) and their 95% confidence intervals (CIs) were obtained to measure the association between the DNA methylation levels and LC. For comparison of clinical characteristics between two or multiple groups, we conducted non-parametric tests including Mann-Whitney U-test and Kruskal-Wallis test. All tests were two-sided, and P < 0.05 was considered to be statistically significant.
Results
The Association Between Aberrant PNPLA2 Methylation in Peripheral Blood and Early-Stage LC
To explore the association between PNPLA2 methylation and LC at an early stage, we conducted a binary logistic regression analysis in a case-control study. When controlling for age, gender and experimental batches, per 10% decrease in methylation level for PNPLA2_CpG_8,10 presented an OR of 1.44 (95% CI: 1.06–1.96, P = 0.018; Table 2). We also found an association between hypermethylation of PNPLA2_CpG_9 and LC with an OR of 0.82 (95% CI: 0.69–0.98, P = 0.029; Table 2). This aberrant methylation of PNPLA2_CpG_8,10 and PNPLA2_CpG_9 was also observed significantly in LC patients at very early stage (stage I) (for PNPLA2_CpG_8,10, OR = 1.42, 95% CI: 1.05–1.93, P = 0.025; for PNPLA2_CpG_9, OR = 0.80, 95% CI: 0.66–0.96, P = 0.017; Table 3). Apart from these, no significant associations were observed between any of the remaining PNPLA2 CpG sites and LC or LC at stage I (P values > 0.05 for all, Tables 2 and 3).
 |
Table 2 The Association Between PNPLA2 Methylation and LC |
 |
Table 3 The Association Between PNPLA2 Methylation and LC at Very Early Stage (Stage I) |
The Association Between Blood-Based PNPLA2 Methylation and Early-Stage LC Stratified by Gender and Age
To eliminate the confounding effects of gender and age, we further evaluated the association between PNPLA2 methylation in peripheral blood and LC by stratified regression analyses. After stratified by gender, per 10% decrease in methylation for PNPLA2_CpG_8,10 conferred an 85% increase on the risk of LC in females (OR =1.85, 95% CI: 1.07–3.22, P = 0.029), but not in males (Table 4). The methylation of PNPLA2_CpG_9 showed a borderline significant association with LC in males (OR =0.75, 95% CI: 0.56–1.00, P = 0.052), but not in females (Table 4). The rest 17 PNPLA2 CpG sites showed no association with LC in either gender (P values > 0.05 for all, Table 4).
 |
Table 4 Gender-Stratified Association Between PNPLA2 Methylation and LC |
In addition, subjects were stratified by the age of 55 years old. In participants less than 55 years old, hypermethylation of PNPLA2_CpG_6 and PNPLA2_CpG_9 was associated with LC (for PNPLA2_CpG_6, OR = 0.77, 95% CI: 0.63–0.95, P = 0.012; for PNPLA2_CpG_9, OR = 0.75, 95% CI: 0.60–0.94, P = 0.011; Table 5). The association between hypomethylation of PNPLA2_CpG_8,10 and LC was enhanced in this younger group with an OR of 2.43 (95% CI: 1.45–4.05, P = 0.001; Table 5). However, none of the 20 measurable CpG sites in PNPLA2 was associated with LC in the subjects older than 55 years old (P values > 0.05 for all, Table 5).
 |
Table 5 Age-Stratified Association Between PNPLA2 Methylation and LC |
The Correlation Between the Level of PNPLA2 Methylation and the Clinical Characteristics of LC
To understand the correlation between methylation pattern of PNPLA2 and the clinical characteristics of LC, the 168 early-stage LC cases were stratified by their clinical information and analyzed by nonparametric tests. Compared to the patients with smaller cancer (tumor length ≤ 1 cm), the LC patients with tumor larger than 1 cm have significantly higher methylation at PNPLA2_CpG_2,3 (median of methylation: 0.15 vs 0.25, P = 0.006; Table 6), which indicated that hypermethylation of PNPLA2_CpG_2,3 might be associated with the progress of LC. Although PNPLA2_CpG_9 and PNPLA2_CpG_20,21 showed significantly different methylation between the stage I and stage II LC patients (Table 6), this result should be taken with caution own to the very limited samples of stage II tumor.
 |
Table 6 The Correlation Between PNPLA2 Methylation and the Clinical Characteristics of LC Patients |
Discussion
In our present case-control study, we measured PNPLA2 DNA methylation level in peripheral blood and found an association with LC in 168 early-stage LC cases and 187 age- and gender-matched healthy controls. PNPLA2 gene is a rate-limiting enzyme catalyzing the first step of intracellular triglycerides hydrolysis in several tissues, and subsequently affects the energy metabolism,10 which is a typical feature of many cancer types.15 Zagani et al found that increased PNPLA2 activity could facilitate cell growth and attenuate apoptosis in NSCLC.16 Intriguingly, PNPLA2 gene is located on chromosome 11p15.5, which was known as a tumor suppressor gene region.17 Loss of PNPLA2 in the somatic tissue was found in 38% of lung cancer and in many other cancer types.18 The mRNA and protein levels of PNPLA2 were lower in four lung adenocarcinoma cells than in normal cells, and the depletion of PNPLA2 may cause triacylglycerol accumulation in lipid droplets and present an aggressive phenotype.19,20
DNA methylation is an early event of cancer. Our findings showed that the changed PNPLA2 methylation level was associated with LC and even with LC at a very early stage (stage I). In our study, the DNA was extracted from the whole blood. The proportion of tumor DNA in blood is minuscule compared with blood cell DNA (ratio about 1:1000),21 and would be very unlikely to change the overall blood methylation values. Thus, the DNA obtained from the whole blood should mostly come from the leukocytes, as the platelet has no nuclear. Other studies have also revealed that the blood leukocyte DNA methylation is associated with LC.22,23 Our previous studies have also disclosed that the breast cancer-associated altered methylation in the blood was mostly originated from the T cells,14,24 which further confirmed that the leukocytes origination for the changed DNA methylation signatures in the whole blood. Unfortunately, there is no fresh blood available in this study to investigate which leukocytes subpopulation is the main contributor for the LC-associated PNPLA2 methylation. So far, there is no direct study to explore the effects of PNPLA2 methylation on its functions. Our target sequence is mainly located at the promoter region, where the changed DNA methylation could regulate the expression of genes.25 Here, we assumed that the changed methylation of PNPLA2 may affect gene functions by modulating its expression and also call for functional studies in the future. The correlation between PNPLA2 methylation and expression should also be investigated when RNA materials are available in further studies. In addition, it would also be interesting to investigate the methylation differences of PNPLA2 among LC cases with different stages.
In gender-stratified analyses, the association of PNPLA2_CpG_8,10 hypomethylation and the risk of LC was observed only in women. On the one hand, it is well documented that inherent DNA methylation differences in peripheral blood between men and women exist in many CpG sites, which can be partly attributed to the difference in circulating sex hormones.26 On the other hand, the evidence showed that female sex hormones including estrogen and its receptor (ER) are vital for lung development and related to the process of LC.27 Both ERα and ERβ exerted a promoting effect on LC via multiple pathways.28,29 Previous studies have identified that women tend to have higher incidence rates of LC, particularly at a younger age, which cannot be explained by smoking habits between gender disparity.30 A comparative study implied that high sensitivity to the tobacco carcinogens in women was another reason for their increased LC morbidity.31 Although tobacco consumption was not available in our study, gender-specific association should be marked.
Unlike our previous study in which the association between hypomethylation of RAPSN and LC was observed in subjects older than 55 years, the present age-stratified data analyses revealed a significant association between aberrant PNPLA2 methylation and the risk of LC in subjects aged ≤ 55 years, highlighting that the effect of changed methylation on LC may be age-related.9 Nevertheless, the sample size of our study is limited. The underlying mechanism of the age-related methylation pattern needs to be further explored in the future studies with large sample size and in a multi-center setting.
To our knowledge, this is the first study to assess the association between PNPLA2 methylation in peripheral blood and LC. Our study recruited more than three hundred participants and 94% LC cases are at a very early stage. However, limited by hospital-based sample collection, information about lifestyle is not available. A prospective cohort and functional studies would be warranted to consolidate the relationship between changed methylation and LC.
Conclusion
In summary, our study disclosed the association between altered PNPLA2 methylation in the peripheral blood and early-stage LC. Also, there are unmet needs for large-scale studies with comprehensive information to verify our findings and even functional experiments to elucidate the molecular mechanism.
Ethics Approval and Informed Consent
The present study was approved by the Ethics Committee of the Shanghai Chest Hospital and Jiangsu Province Hospital of Chinese Medicine in China. Written informed consents were obtained from all the participants.
Acknowledgments
We are grateful to Feifei Di, Jin Zhang and Jiao Yang from Nanjing TANTICA Biotechnology Co. Ltd for their contribution to the sample processing.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Nanjing Social Supporting Department and Social Supporting Ministry of Jiangsu Province (Grant No. 20182020) and by the Nanjing TANTICA Co. Ltd with grant number 2018LC01.1.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Wang N, Mengersen K, Tong S, Kimlin M, Zhou M, Hu W. Global, regional, and national burden of lung cancer and its attributable risk factors, 1990 to 2017. Cancer. 2020;126(18):4220–4234. doi:10.1002/cncr.33078
3. Chansky K, Detterbeck FC, Nicholson AG, et al. The IASLC lung cancer staging project: external validation of the revision of the TNM stage groupings in the eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2017;12(7):1109–1121. doi:10.1016/j.jtho.2017.04.011
4. National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409. doi:10.1056/NEJMoa1102873
5. Shen H. Low-dose CT for lung cancer screening: opportunities and challenges. Front Med. 2018;12(1):116–121. doi:10.1007/s11684-017-0600-1
6. Greenberg MVC, Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20(10):590–607. doi:10.1038/s41580-019-0159-6
7. Costa-Pinheiro P, Montezuma D, Henrique R, Jeronimo C. Diagnostic and prognostic epigenetic biomarkers in cancer. Epigenomics. 2015;7(6):1003–1015. doi:10.2217/epi.15.56
8. Baglietto L, Ponzi E, Haycock P, et al. DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. Int J Cancer. 2017;140(1):50–61. doi:10.1002/ijc.30431
9. Qiao R, Di F, Wang J, et al. The association between RAPSN methylation in peripheral blood and early stage lung cancer detected in case-control cohort. Cancer Manag Res. 2020;12:11063–11075. doi:10.2147/CMAR.S275321
10. Nielsen TS, Jessen N, Jorgensen JO, Moller N, Lund S. Dissecting adipose tissue lipolysis: molecular regulation and implications for metabolic disease. J Mol Endocrinol. 2014;52(3):R199–R222. doi:10.1530/JME-13-0277
11. Vegliante R, Di Leo L, Ciccarone F, Ciriolo MR. Hints on ATGL implications in cancer: beyond bioenergetic clues. Cell Death Dis. 2018;9(3):316. doi:10.1038/s41419-018-0345-z
12. Liu M, Yu X, Lin L, et al. ATGL promotes the proliferation of hepatocellular carcinoma cells via the p-AKT signaling pathway. J Biochem Mol Toxicol. 2019;33(11):e22391. doi:10.1002/jbt.22391
13. Yang X, Huang Y, Sun C, Li J. Maternal prenatal folic acid supplementation programs offspring lipid metabolism by aberrant DNA methylation in hepatic ATGL and adipose LPL in rats. Nutrients. 2017;9(9):935. doi:10.3390/nu9090935
14. Yang R, Stocker S, Schott S, et al. The association between breast cancer and S100P methylation in peripheral blood by multicenter case-control studies. Carcinogenesis. 2017;38(3):312–320. doi:10.1093/carcin/bgx004
15. Chen X, Qian Y, Wu S. The Warburg effect: evolving interpretations of an established concept. Free Radic Biol Med. 2015;79:253–263. doi:10.1016/j.freeradbiomed.2014.08.027
16. Zagani R, El-Assaad W, Gamache I, Teodoro JG. Inhibition of adipose triglyceride lipase (ATGL) by the putative tumor suppressor G0S2 or a small molecule inhibitor attenuates the growth of cancer cells. Oncotarget. 2015;6(29):28282–28295. doi:10.18632/oncotarget.5061
17. Bepler G, Garcia-Blanco MA. Three tumor-suppressor regions on chromosome 11p identified by high-resolution deletion mapping in human non-small-cell lung cancer. Proc Natl Acad Sci U S A. 1994;91(12):5513–5517. doi:10.1073/pnas.91.12.5513
18. Al-Zoughbi W, Pichler M, Gorkiewicz G, et al. Loss of adipose triglyceride lipase is associated with human cancer and induces mouse pulmonary neoplasia. Oncotarget. 2016;7(23):33832–33840. doi:10.18632/oncotarget.9418
19. Tomin T, Fritz K, Gindlhuber J, et al. Deletion of adipose triglyceride lipase links triacylglycerol accumulation to a more-aggressive phenotype in A549 lung carcinoma cells. J Proteome Res. 2018;17(4):1415–1425. doi:10.1021/acs.jproteome.7b00782
20. Zhou Q, Sun Y. Circular RNA cMras suppresses the progression of lung adenocarcinoma through ABHD5/ATGL axis using NF-kappaB signaling pathway. Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2020.3709
21. Hoque MO, Feng Q, Toure P, et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol. 2006;24(26):4262–4269. doi:10.1200/JCO.2005.01.3516
22. Woo HD, Kim J. Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis. PLoS One. 2012;7(4):e34615. doi:10.1371/journal.pone.0034615
23. Zhao N, Ruan M, Koestler DC, et al. Epigenome-wide scan identifies differentially methylated regions for lung cancer using pre-diagnostic peripheral blood. Epigenetics. 2021:1–13. doi:10.1080/15592294.2021.1923615
24. Yang R, Pfutze K, Zucknick M, et al. DNA methylation array analyses identified breast cancer-associated HYAL2 methylation in peripheral blood. Int J Cancer. 2015;136(8):1845–1855. doi:10.1002/ijc.29205
25. Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392(10149):777–786. doi:10.1016/S0140-6736(18)31268-6
26. Singmann P, Shem-Tov D, Wahl S, et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenetics Chromatin. 2015;8:43. doi:10.1186/s13072-015-0035-3
27. Stabile LP, Davis AL, Gubish CT, et al. Human non-small cell lung tumors and cells derived from normal lung express both estrogen receptor alpha and beta and show biological responses to estrogen. Cancer Res. 2002;62(7):2141–2150.
28. Xu L, Liao Y, Tang H, Zhang C, Liu Z. Advances of targeted therapy based on estrogen receptor signaling pathway in lung cancer. Zhongguo Fei Ai Za Zhi. 2011;14(9):727–732. doi:10.3779/j.issn.1009-3419.2011.09.07
29. He M, Yu W, Chang C, et al. Estrogen receptor alpha promotes lung cancer cell invasion via increase of and cross-talk with infiltrated macrophages through the CCL2/CCR2/MMP9 and CXCL12/CXCR4 signaling pathways. Mol Oncol. 2020;14(8):1779–1799. doi:10.1002/1878-0261.12701
30. Jemal A, Miller KD, Ma J, et al. Higher lung cancer incidence in young women than young men in the United States. N Engl J Med. 2018;378(21):1999–2009. doi:10.1056/NEJMoa1715907
31. International Early Lung Cancer Action Program Investigators. Women’s susceptibility to tobacco carcinogens and survival after diagnosis of lung cancer. JAMA. 2006;296(2):180–184. doi:10.1001/jama.296.2.180
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
