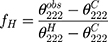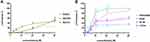Back to Journals » Drug Design, Development and Therapy » Volume 15
The Antimicrobial Peptide Melectin Shows Both Antimicrobial and Antitumor Activity via Membrane Interference and DNA Binding
Authors Liang X, Yan J, Lu Y, Liu S, Chai X
Received 30 October 2020
Accepted for publication 5 February 2021
Published 19 March 2021 Volume 2021:15 Pages 1261—1273
DOI https://doi.org/10.2147/DDDT.S288219
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Anastasios Lymperopoulos
Xiaolei Liang,1,* Jiexi Yan,2,* Yingwei Lu,3 Shan Liu,4 Xiaojing Chai5
1Key Laboratory for Gynecologic Oncology Gansu Province, Department of Obstetrics and Gynecology, The First Hospital of Lanzhou University, Lanzhou, People’s Republic of China; 2The Precision Medicine Laboratory, The First Hospital of Lanzhou University, Lanzhou, People’s Republic of China; 3Gansu Provincial Center for Disease Control and Prevention, Lanzhou, People’s Republic of China; 4The First Clinical Medicine School, Lanzhou University, Lanzhou, People’s Republic of China; 5The Key Laboratory, The First Hospital of Lanzhou University, Lanzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaojing Chai
The Key Laboratory, The First Hospital of Lanzhou University, No. 1, Donggang Road, Lanzhou, People’s Republic of China
Email [email protected]
Purpose: Increasingly complex diseases require novel drugs for their treatment. Antimicrobial peptides (AMPs) are promising candidate treatments due to their broad existence and special characteristics. However, the current understanding of AMPs is not sufficient to allow them to be produced commercially for clinical use.
Materials and Methods: Melectin, from the venom of the cleptoparasitic bee Melecta albifrons, does not exhibit sequence homology with other wasp venom peptides. To investigate this more deeply, we explored the antibacterial and antitumor activities of Melectin and related mechanisms.
Results: Our results demonstrate that Melectin possesses antimicrobial properties against standard sensitive/clinical drug-resistant bacteria strains as well as antitumor activity. It has an α-helix form and exhibits moderate cytotoxicity. Its action mechanisms are involved with membrane interfering and DNA binding. The membrane interfering effect was distinct between different phospholipid compositions.
Conclusion: We found that Melectin may serve as a new potential template in the battle against multidrug resistance, and our study indicated that there are promising prospects for medically applicable drugs based on AMPs.
Keywords: antimicrobial peptide, melectin, antibacterial, antitumor, action mechanism
Introduction
Disease is a serious threat to quality of life. Researchers are working to prevent or treat disease and to prolong and improve quality of life. As our understanding of disease deepens and science and technology continue to develop, more and more novel pre-drugs, distinct from traditional drugs, are emerging, with some finding clinical application.1 Among these, antimicrobial peptides (AMPs) in particular are a hot spot for research due to their special characteristics. AMPs are the first line of defense in the innate immune system in many organisms.2 They are normally composed of fewer than 100 amino acids and have diverse structures.3 AMPs exhibit broad-spectrum bioactivity against various microbes and regulate innate immunity.4 Due to their various bioactivities and special mechanisms of action, AMPs are a promising alternative or supplement to traditional drugs.
Approximately three thousand AMPs, including synthetic peptides from six kingdoms, are recorded in the AMP database.5 Of these, the AMPs derived from insects form a notable group. Insects are distinctively adapted to complex living environments. They have developed amazing defense systems using hemolymphs and venom to subdue prey and defend themselves against predators. Melectin was first found in the venom of the cleptoparasitic bee Melecta albifrons, and it has been noted that it does not exhibit sequence similarity with other wasp venom peptides.6 A previous study found that Melectin possesses antimicrobial activity against Gram positive and Gram negative strains of bacteria. It has also been found to degranulate peritoneal mast cells in rats.7
To investigate this more deeply, we explored the antibacterial and antitumor activities of Melectin and related mechanisms. We found that Melectin not only possesses antimicrobial properties against standard sensitive/clinical drug-resistant bacteria strains but also has antitumor activity, indicating that it has great potential as an AMP template for future applications.
Materials and Methods
Peptides
Melectin was purchased from company (GL Biochem, Shanghai, China). The homogeneity was greater than 95%. The atomic mass of peptide was confirmed by electrospray ionization mass spectrometry.
Bacterial Strains and Cell Lines
The standard bacterial strains utilized in this study were purchased from American type culture collection (ATCC). Drug-resistant bacterial strains were originally isolated from clinical samples and preserved in the Culture Collection Center at School of Basic Medical Sciences of Lanzhou University (Lanzhou, China). All strains of bacteria were cultured following ATCC’s instructions. The cell lines used in this study were gifted from the basic medical sciences school of Lanzhou University. The leukemia cell lines were K562, K562/ADM, HL-60, THP-1, and Jurkat, and the noncancerous cell lines were HUVEC, NIH3T3, and HEK293. All of the cell lines except NIH3T3 were cultured in RP1640 medium with 10% FBS at 37°C in 5% CO2. The NIH3T3 cells were cultured in DMEM medium with 10% FBS at 37°C in 5% CO2. The experimental protocols were approved by the Ethics Committee of the First Hospital of Lanzhou University (LDYYLL2015–0007). The use of cell lines was approved by the institutional review board of the School of Basic Medical Sciences and the First Hospital of Lanzhou University.
Antibacterial Activity of Melectin
Radial Diffusion Assay
Bacteria were cultured to an optical density of 0.5 at 600 nm; 100 mL MH agar (Hopebio, Qingdao, China) was previously autoclaved and cooled to a suitable temperature. Then 1 mL bacteria was added to the agar, and the suspension was given a rapid dispersion in the agar plate.8 After the agar formed a layer about 5 mm deep, it was punched with a gel punch 3 mm in diameter. After this, different concentrations of Melectin were added to every well, and the agar plates were incubated for 24 h at 37°C. Sterile water was used as a control. The clear zone surrounding each well was photographed.
Determination of Minimum Inhibition Concentrations of Melectin Against Drug-Sensitive/Resistant Bacteria
The minimum inhibition concentrations (MICs) of Melectin against drug-sensitive or drug-resistant bacteria were measured following the broth microdilution procedure, the method used by the Clinical and Laboratory Standards Institute.9 Briefly, Melectin was serially diluted two-fold in MH medium, 100 µL bacterial suspension was added to a 96-well microtiter plate, and 100 µL concentrations of Melectin were added. The MH broth served as control. The MICs were identified by the absence of turbidity after incubation for 24 h at 37°C. All of the tests were performed in triplicate.
Killing Kinetics of Melectin
To establish an understanding of the effects of Melectin against bacteria over time, a killing kinetics assay was carried out following a modification of a method described previously.10 That is, 100 µL bacterial suspension was treated with 100 µL Melectin with different concentrations in a 96-well plate at 37°C. At the defined time point, one of the 96-well plates was chosen, and the absorbance at 450 nm was measured using a microplate reader (Bio-Rad 680, CA, USA).
Propidium Iodide Uptake Detection
The integrity of the bacterial membrane treated with Melectin was reflected by the propidium Iodide (PI) uptake.11 In brief, E. coli bacteria were treated with 10 µM Melectin for 30 min at 37°C. Then a PI solution (20 μg/mL) was added to stain for 30 min, shielded from light and at room temperature. The bacteria were centrifuged and washed three times with PBS buffer and dropped on a glass slide. The integrity of the bacterial membrane was photographed using a confocal laser scanning microscope (CLSM, Leica SP2, Germany).
Outer and Inner Membrane Permeability
The outer membrane (OM) permeability induced by Melectin in E. coli (ATCC 25922) was recorded using the fluorescent probe N-phenyl-1-naphthylamine (NPN). The inner membrane (IM) permeation was reflected by the cytoplasmic β-galactosidase release activity of E. coli ML-35.12,13 The assays were conducted following methods previously described with slight modification.10,14 In brief, E. coli ATCC 25922 (OD600=0.5±0.02) was centrifuged and suspended in 5 mM HEPES. Then 100 µL E. coli was mixed with 50 µL Melectin (final concentrations: 4 µM, 8 µM, and 16 µM) and 50 µL NPN (final concentration: 10 µM). The negative control was 0.5% NaCl. Then fluorescence was recorded (λexc=350 nm, λemi=420 nm). Next, 100 µL E. coli (ML-35) was mixed with 90 µL Melectin (final concentrations: 4 µM, 8 µM, and 16 µM) and 10 µL o-Nitrophenyl-β-D-galactoside (ONPG,30 mM) in a 96-well plate. The controls were 0.5% NaCl and Triton X-100. The production of o-nitrophenol over time was determined by monitoring the changes in absorbance at 420 nm.
Calcein Release from Large Unilamellar Vesicles
The ability of Melectin to cause content leakage from liposomes was assessed against different large unilamellar vesicles (LUVs). LUVs composed of phosphatidylcholine and cholesterol (9:1, wt:wt) (Avanti Polar, Alabama, USA) were employed to mimic a neutrally charged normal cell membrane, and those composed of DPPG/DPPE (7:3, wt:wt) were used to mimic a negatively charged bacterial membrane. The preparation of LUVs containing calcein followed a previous work.15 The calcein release from different LUVs was monitored by using a plate reader (Tecan Infinite M200 Pro, Mannedorf, Switzerland) by recording the fluorescence intensity (λexc=490 nm, λemi=520 nm). The positive control was 2% Triton X-100 in Tris buffer, and PBS buffer was utilized as the negative control. The following formula was used to calculate the percentage of dye leakage caused by the Melectin: (F-F0)/(Ft-F0)×100%, where F is the fluorescence intensity after Melectin addition, F0 is fluorescence intensity with PBS buffer, and Ft is fluorescence intensity with Triton X-100.
Transmission Electron Microscopy (TEM) Scanning
The LUVs used for TEM analyses were prepared as previously described.16 Different LUVs treated with 50 μg/mL Melectin or not were deposited on copper grids coated with carbon and dried naturally. To stain the grids, 2% phosphotungstic acid was used for 1 min. After the additional phosphotungstic acid was soaked up, electron micrographs were recorded with an accelerating voltage of 120 kV (JEM-1230; Japan).
DNA Binding Activity of Melectin
A gel retardation experiment was used to reflect Melectin DNA binding activity, as described previously.17 In brief, plasmid DNA (pBR322, 300 ng) was mixed with Melectin (the final concentrations were 0.5, 1, 2, 4, 8, 16, 32, and 64 μM) in 30 µL buffer (10 mM Tris-HCl, 1 mM EDTA buffer, pH 8.0) and reacted for 30 min at room temperature. After that, 20 µL mixture was used for gel electrophoresis (agarose concentration was 1.5%) and DNA migration was marked by ethidium bromide.
Antitumor Activity of Melectin
Cell Proliferation Assay
The antitumor activity of Melectin was studied using an MTT assay, following a previous report:18 1×104 cells/well were co-incubated with different concentrations of Melectin or PBS buffer for 24 h. After adding 10 µL MTT (5 mg/mL) solution for 4 h incubation, 100 µL SDS-isobutanol-HCl solution (10% SDS, 5% isobutanol, and 12 μM HCl) was added to the wells and reacted overnight. Then the microplate was detected under a microplate reader (Bio-Rad 680, CA, USA) and absorbance was recorded at 570 nm.
Hemolytic Activity of Melectin
The hemolytic activity of Melectin was detected according to a method described previously.19 Peripheral blood from volunteers was freshly collected in a heparinized tube. All volunteers had signed an informed consent form, and the process was approved by the Ethics Committee of the First Hospital of Lanzhou University (LDYYLL2019-79). After 800 × g centrifugation for 10 min, the pellet was washed with PBS and re-suspended (final erythrocyte concentration was 8%). Then 100 µL suspension in a microtiter plate was co-incubated with Melectin for 60 min at 37°C. The positive and negative controls were Triton-X 100 (0.2%) and PBS buffer, respectively. After 1000 × g centrifugation for 10 min, the hemoglobin release was measured at 490 nm.
Lactate Dehydrogenase (LDH) Assay
The cytotoxicity of Melectin was reflected by LDH leakage. Briefly, 1 × 104/well cells were treated with different concentrations of Melectin for 4 h at 37°C. Then the LDH release was conducted according to the manufacturer’s instructions (Cytotoxicity Detection KitPLUS [LDH], Roche, Basel, Switzerland). After microplate centrifugation, 100 µL medium was transferred to another 96 well plate and LDH reaction mixture was added to react for 15 min. After that, the absorbance at 490 nm was detected using a microplate reader.
Scanning Electron Microscope Examination of Tumor Cells Treated with Melectin
Tumor cells were seeded in a six-well microplate. Then 10 µM Melectin was added and incubated for 30 min. The cells were harvested and washed with PBS buffer. Fixation and dehydration were done according to a method described previously.16 In brief, the cell pellets were suspended in 3% glutaraldehyde solution and soaked in 2.5% tannic acid for 2 days. Then the cells were treated with 2% osmium tetroxide and dehydrated in ethanol. Finally, the cells were dried and coated with gold. The membrane change was recorded using a scanning electron microscope (SEM) (JSM-6380Lv, Japan).
Circular Dichroism Spectrum
A circular dichroism (CD) spectrum was utilized to reflect the secondary structure of Melectin.17 Under a nitrogen flush in 1 mm path length, an Olis DSM 1000 CD spectrophotometer (USA) was operated at room temperature. The spectra of Melectin solution (dissolved in 50% TFE and 10 mM PBS buffer) were recorded between 195 nm and 240 nm. The final concentration of Melectin was 50 μM. The percentage of α-helical content was calculated using the following formula:
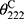 , which represents the complete random coil ellipticity, is equal to 640.
, which represents the complete random coil ellipticity, is equal to 640. 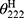 represents the mean ellipticity of complete helical conformation, and it is calculated using this formula:
represents the mean ellipticity of complete helical conformation, and it is calculated using this formula:
Where n is the chain length in residues and 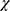 is the number of non H-bonded carbonyl groups in the peptides, as proposed by Rohl and Baldwin.20
is the number of non H-bonded carbonyl groups in the peptides, as proposed by Rohl and Baldwin.20
Statistical Analyses
At least three independent experiments were conducted. Statistical analyses were performed using GraphPad Prism software. Data from the experiments are expressed as means ± SEM and analyzed with Student’s t-test or one-way analysis of variance. Statistical significance was set to p<0.05.
Results
Peptide
The primary sequence and related biophysical parameters of Melectin are given in Table 1. Melectin consists of 18 amino acids and confers a positive net charge of 5. The mean hydrophobicity (H) and hydrophobic moment of Melectin were calculated according to the consensus scale proposed by Eisenberg and McLachlan.21 A prediction was made regarding the secondary structure of Melectin, and the results showed that Melectin adopted an α-helix structure based on the software supplied by http://heliquest.ipmc.cnrs.fr/.
 |
Table 1 Amino Sequence, Molecular Weight and Biophysical Parameters of Melectin |
Melectin Shows Potent Antibacterial Activity
Antibacterial Activity of Melectin as Assessed via the Radio Diffusion and MIC Assays
Radial diffusion assay was used to screen the antibacterial activity of Melectin. As shown in Figure 1, Melectin showed remarkable antibacterial activity against E. coli and S. aureus. It induced a markedly clear zone that was larger than that of the control. The diameter of the inhibition zone increased with an increase in peptide concentration, indicating that the antimicrobial activity of Melectin acted in a concentration-dependent way. The MICs of Melectin were determined using the broth microdilution method with different bacteria strains, including drug-sensitive and drug-resistant bacteria. The MICs of the drug-resistant bacteria to several antibiotics had been detected in a previous study.14 According to our results, Melectin showed excellent antibacterial activity against both of these bacterial strains (Table 2).
 |
Table 2 The Minimal Inhibition Concentrations (MICs) of Melectin Against Drug-Sensitive and Drug-Resistant Bacterial Strains |
Killing Kinetics of Melectin
To obtain a detailed understanding of the antibacterial activity of Melectin, time-killing kinetic assays were carried out against E. coli and S. aureus to reflect the bacteria changes over time after Melectin treatment. As shown in Figure 2, Melectin delivered a rapid killing effect against both E. coli and S. aureus, although the details were different between them. For E. coli, Melectin only slightly inhibited bacterial growth at a concentration of 4 μM. When the concentration reached 8 μM, its killing effect was obviously strengthened, and it was effective within 50 min. Compared to E. coli, the killing kinetics of Melectin against S. aureus came into effect within 50 min at a concentration of 4 μM. The bacteria killing rate was similar between the two bacterial strains.
PI Uptake Assay
It has been reported that most AMPs can disrupt the target cell membrane. To identify the effects of Melectin on the bacterial membrane, PI, as a DNA-staining fluorescent probe, was used in this study. It was found that the majority of E. coli cells turned red after treatment with Melectin for 30 min (Figure 3). This result indicates that Melectin induces PI influx after destabilizing or disturbing the integrity of the membrane. That is, it affected the membrane as most AMPs do.
Effects of Melectin on the Outer and Inner Membrane of Bacteria
According to the PI uptake assay, we concluded that Melectin disrupted the membrane of E. coli. The membrane of E. coli is constructed in two layers: the OM and the IM. To produce a detailed understanding of how Melectin affected in E. coli, OM and IM permeability were both measured. NPN is a hydrophobic fluorescent probe, and its fluorescence is powerless when it is in an aqueous environment; however, it increases in brightness in the hydrophobic core of the bacterial membrane.22 Cytoplasmic β-galactosidase from E. coli can be released when the IM is altered or compromised. It can react with ONPG and produce ortho-nitrophenol.23 Thus, NPN and ONPG were employed to reflect the membrane situation of E. coli after Melectin treatment. As shown in Figure 4A, the fluorescence of NPN increased sharply in a dose-dependent way when it was quickly treated with Melectin, and it reached a maximum at 4 min. The gradual release of β-galactosidase from E. coli cells displayed a steady state at the time point of 60 min, when the concentration of Melectin was 16 μM (Figure 4B). The release manner of the β-galactosidase was also dose dependent.
Effects of Melectin on LUVs Composed from Different Phospholipids
LUVs with different phospholipid components were used to intuitively reflect the interaction between Melectin and the distinct membranes. LUVs constructed by negatively charged DPPG/DPPE (7:3, wt:wt) were used to mimic E. coli membrane composition, and LUVs composed of neutrally charged PC and cholesterol (9:1, wt:wt) were used to mimic the normal cell membrane. As shown in Figure 5, Melectin induced an obvious calcein leakage as soon as it interacted with LUVs and this ability was concentration dependent. However, this ability was significantly different between these two kinds of LUVs. The calcein released from the negatively charged LUVs was much more than that from the neutrally charged LUVs. This result indicates that the interaction activity of Melectin differs between bacterial and normal cell membranes.
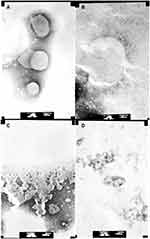
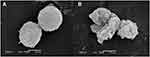
At the same time, different LUVs were incubated with 50 μg/mL Melectin, and TEM was employed to visualize their surface changes. Compared to the group treated with PBS buffer (Figure 6A), the membranes of neutral LUVs were only slightly influenced, the margin of membrane could still be seen and its structure was relatively intact (Figure 6B). However, the negatively charged LUVs changed thoroughly after treatment. The membrane was partial, and the margin was unclear. Their structures were difficult to observe, and all of the liposomes seemed to break into pieces and attach to the copper grids (Figure 6C and D). This result indicates that Melectin has different effects when acting against liposomes with different phospholipid components.
Melectin Interacts with DNA
Our results indicate that Melectin exhibits activity against bacterial membranes and its mimetic liposomes. To investigate whether Melectin has other targets apart from bacterial membranes, plasmid DNA was used to examine the interactions between Melectin and DNA. According to its electrophoretic mobility on an agarose gel, the migration of plasmid DNA appeared remarkably different after treatment with Melectin. As shown in Figure 7, the plasmid DNA that migrated into the gel was almost the same as the non-treated DNA when Melectin concentration was under 2 μM. When the concentration reached 16 μM, most of the plasmid DNA remained at the origin, and complete DNA retardation was seen. This result indicates that Melectin binds with plasmid DNA, in addition to its interaction with bacterial membranes, and this effect was also concentration dependent.
Melectin Shows Potent Antitumor Activity
Melectin Exhibits Proliferation Inhibition Activity Against Leukemia Cell Lines
To the best of our knowledge, many AMPs do not possess only one bio-function. To fully understand Melectin, its antitumor activity was also explored. A panel of leukemia cell lines was used in this assay. As shown in Figure 8, Melectin obviously inhibited the proliferation of tested leukemia cell lines at a concentration of 15 μM, particularly against the THP-1 and HL-60 cell lines. For the normal cell lines used in this study, although Melectin exhibited a slight inhibition effect, the IC50 concentrations were higher than that of tumor cells. Both of these effects were concentration dependent. The results of proliferation inhibition showed that Melectin possessed antitumor activity with moderate selection.
Hemolytic and Cytotoxicity Activity of Melectin
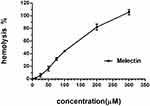
The cytotoxicity of Melectin was evaluated using a hemolytic and LDH release assay. As shown in Figure 9, Melectin showed remarkable hemolysis activity with increasing concentration. It induced 100% hemolysis at the detected concentration of 300 μM. However, this hemolytic ability was limited at concentrations under 50 μM, which only caused less than 20% erythrocyte lysis. In addition to this, the potential cytotoxicity of Melectin was proven by the cytoplasmic enzyme LDH release. As shown in Figure 10, Melectin induced LDH release in a concentration-dependent manner. This was observed both in leukemia cell lines and normal cell lines. However, normal cell lines exhibited obviously lower LDH release. The results of LDH release were similar to those of our previous hemolytic assays, showing that the selectivity of Melectin was moderate.
Melectin Disturbs Tumor Cell Membranes
The effects of Melectin against tumor cell membranes were also investigated via SEM. After treatment with Melectin, significant changes in morphology were observed (Figure 11). Untreated leukemia cells showed an integrated and smooth cell membrane. Conversely, the membrane of Melectin treated leukemia cells was destroyed and became full of bumps and pores. This phenomenon was consistent with the effects of Melectin against the bacterial membrane. This proved that Melectin acted on the tumor cell membranes, just as it did on bacterial membranes, both in a membrane-targeting manner.
Secondary Structure Analyses of Melectin
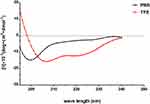
AMPs possess a variety of structures, including α-helix, β-sheet, and random coil. Their biological functions are closely related to their secondary structures. To obtain a deep understanding of Melectin, its second structure was evaluated by CD spectroscopy. It exhibited a random coil structure in PBS buffer, mimicking a benign condition. Additionally, Melectin also presented negative bands at wavelengths of 208 nm and 222 nm when in a 50% TFE environment, which mimics the membrane condition (Figure 12). This indicates that Melectin adopts a typical α-helical structure in a membrane-mimicking environment. We calculated the α-helix contents of Melectin in these two environments, and the results are shown in Table 3.
 |
Table 3 Circular Dichroism Spectrum Data of Melectin in Different Environments |
Discussion
AMPs have attracted increasing attention as multidrug-resistant bacteria continue to emerge. As an AMP isolated from the venom of bees, previous studies have shown that Melectin exhibits excellent antimicrobial activity against sensitive bacteria. At the same time, it can degranulate peritoneal mast cells from rats.24 However, more information on Melectin is needed before it can be seen as a potent treatment candidate. Does it also affect drug-resistant bacteria? What is the mechanism of its action? Does it have any other bioactivity? These problems led us to try to understand this potent AMP template. In this study, we broadened the focus scope of Melectin by investigating its activity against multidrug-resistant bacteria and tumor cells. We likewise explored its possible mechanisms of action.
We found that Melectin showed antibacterial activity not only against sensitive Gram negative and Gram positive bacteria but also against drug-resistant bacteria. This activity was notably time and concentration dependent. A PI uptake assay and the IM/OM assay indicated that Melectin affects bacteria by interfering with the membrane. To present intuitive proof of this, LUVs composed of phospholipids were employed. Our results showed that Melectin affects the lipid membrane, with variations between different phospholipid compositions. It showed a greater affinity to LUVs constructed with negative phospholipids, which could be one reason for its selectivity against normal cells.
According to our results, bacterial cell membranes are a target of Melectin. This result is in accord with previous studies on AMPs.25 However, we still wanted to know whether the cell membrane was the only target of Melectin. We investigated the DNA-binding activity of Melectin with plasmid DNA, and the results showed that Melectin also bound with plasmid DNA. This activity is quite different from another representative venom AMP, magainin. In an earlier study, we found that magainin did not show any DNA binding activity under the same experimental conditions.10 This indicates that DNA might be another target of Melectin apart from the bacterial cell membrane.
We also investigated the antitumor activity of Melectin. It exhibited moderate activity against all of the leukemia cell lines, although it had somewhat different effects on different cell types. However, the cytotoxicity of Melectin is a disadvantage. In subsequent studies, we will pursue additional chemical strategies to modify the primary structure of Melectin to improve its bioactivity and decrease its cytotoxicity. Our results also indicated that Melectin disturbs tumor cell membranes. This finding is similar to the action mode of Melectin in bacteria.
AMPs normally appear in three types of conformation: α-helix, β-sheet, and random coil.21 The α-helix is the most common conformation, and it is essential for the biological function of AMPs. Based on the secondary structure data, Melectin adopted a well-defined α-helix conformation in a membrane-mimicking environment. Combining all of our experimental results, we could speculate that Melectin functions against bacteria via two main mechanisms. First, it binds with bacterial cell membranes through electrostatic attraction and forms pores, leading to bacterial cell content release and death. Second, it directly passes through the bacterial membrane and interacts with target DNA molecules that might consequently affect bacterial growth or metabolism.
Conclusions
Melectin not only possesses antimicrobial activity against sensitive and resistant bacteria but also shows antitumor activity. It exhibits moderate cytotoxicity. Its action mechanisms are related to both membrane perturbation and DNA interaction. This study builds on previous work and suggests that Melectin may serve as a new template in the battle against multidrug resistance. Our results reflect a promising outlook for finding medically applicable drugs through AMPs.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (No. 81960278 and 81601351), the Outstanding Youth Funds of Science and Technology Department of Gansu Province (No. 20JR5RA371), the Natural Science Foundation of Gansu Province (No. 20JR10RA678), the Research Funds from Health Commission of Gansu Province (No. GSWSKY-2019-87), the Fundamental Research Funds for the Central Universities (No. lzujbky-2020-kb28), and the Natural Science Foundation of the First Hospital of Lanzhou University (No. ldyyyn2018-53, ldyyyn2018-52).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bashraheel SS, Domling A, Goda SK. Update on targeted cancer therapies, single or in combination, and their fine tuning for precision medicine. Biomed Pharmacother. 2020;125:110009.
2. Zandsalimi F, Talaei S, Noormohammad Ahari M, et al. Antimicrobial peptides: a promising strategy for lung cancer drug discovery? Expert Opin Drug Discov. 2020;15:12. doi:10.1080/17460441.2020.1791080
3. Wang JJ, Dou X, Song J, et al. Antimicrobial peptides: promising alternatives in the post feeding antibiotic era. Med Res Rev. 2019;39(3):831–859. doi:10.1002/med.21542
4. Patel S, Akhtar N. Antimicrobial peptides (AMPs): the quintessential ‘offense and defense’ molecules are more than antimicrobials. Biomed Pharmacother. 2017;95:1276–1283. doi:10.1016/j.biopha.2017.09.042
5. Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44(D1):D1087–1093. doi:10.1093/nar/gkv1278
6. Cerovsky V, Hovorka O, Cvacka J, et al. Melectin: a novel antimicrobial peptide from the venom of the cleptoparasitic bee Melecta albifrons. ChemBioChem. 2008;9(17):2815–2821. doi:10.1002/cbic.200800476
7. Niederhafner P, Bednárová L, Buděšínský M, et al. Melectin MAPs: the influence of dendrimerization on antimicrobial and hemolytic activity. Amino Acids. 2010;39(5):1553–1561. doi:10.1007/s00726-010-0626-z
8. Takemura H, Kaku M, Kohno S, et al. Evaluation of susceptibility of gram-positive and -negative bacteria to human defensins by using radial diffusion assay. Antimicrob Agents Chemother. 1996;40(10):2280–2284. doi:10.1128/AAC.40.10.2280
9. CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically CLSI Approved Standard - Seventh Edition CLSI Document M7-A72006. CLSI; 2006.
10. Yan J, Wang K, Dang W, et al. Two hits are better than one: membrane-active and DNA binding-related double-action mechanism of NK-18, a novel antimicrobial peptide derived from mammalian NK-lysin. Antimicrob Agents Chemother. 2013;57(1):220–228. doi:10.1128/AAC.01619-12
11. Wang K, Dang W, Yan J, et al. Membrane perturbation action mode and structure-activity relationships of Protonectin, a novel antimicrobial peptide from the venom of the neotropical social wasp Agelaia pallipes pallipes. Antimicrob Agents Chemother. 2013;57(10):4632–4639. doi:10.1128/AAC.02311-12
12. Wang B, Navath RS, Menjoge AR, et al. Inhibition of bacterial growth and intramniotic infection in a guinea pig model of chorioamnionitis using PAMAM dendrimers. Int J Pharm. 2010;395(1–2):298–308. doi:10.1016/j.ijpharm.2010.05.030
13. Lee D, Je J. Gallic acid-grafted-chitosan inhibits foodborne pathogens by a membrane damage mechanism. J Agric Food Chem. 2013;61(26):6574–6579. doi:10.1021/jf401254g
14. Xie J, Gou Y, Zhao Q, et al. Antimicrobial activities and membrane-active mechanism of CPF-C1 against multidrug-resistant bacteria, a novel antimicrobial peptide derived from skin secretions of the tetraploid frog Xenopus clivii. J Pept Sci. 2014;20(11):876–884. doi:10.1002/psc.2679
15. Zhu W, Song YM, Park Y, et al. Substitution of the leucine zipper sequence in melittin with peptoid residues affects self-association, cell selectivity, and mode of action. Biochim Biophys Acta. 2007;1768(6):1506–1517. doi:10.1016/j.bbamem.2007.03.010
16. Yan J, Liang X, Bai C, et al. NK-18, a promising antimicrobial peptide: anti-multidrug resistant leukemia cells and LPS neutralizing properties. Biochimie. 2018;147:143–152. doi:10.1016/j.biochi.2018.02.001
17. Yan J, Liang X, Liu C, et al. Influence of proline substitution on the bioactivity of mammalian-derived antimicrobial peptide NK-2. Probiotics Antimicrob Proteins. 2018;10:118–127. doi:10.1007/s12602-017-9335-1
18. Yan JX, Wang K-R, Chen R, et al. Membrane active antitumor activity of NK-18, a mammalian NK-lysin-derived cationic antimicrobial peptide. Biochimie. 2012;94(1):184–191. doi:10.1016/j.biochi.2011.10.005
19. Wang KR, Zhang B-Z, Zhang W, et al. Antitumor effects, cell selectivity and structure-activity relationship of a novel antimicrobial peptide polybia-MPI. Peptides. 2008;29(6):963–968. doi:10.1016/j.peptides.2008.01.015
20. Rohl CA, Baldwin RL. Deciphering rules of helix stability in peptides. Methods Enzymol. 1998;295:1–26.
21. Eisenberg D, McLachlan AD. Solvation energy in protein folding and binding. Nature. 1986;319(6050):199–203. doi:10.1038/319199a0
22. Loh B, Grant C, Hancock RE. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1984;26(4):546–551. doi:10.1128/AAC.26.4.546
23. Falla TJ, Hancock RE. Improved activity of a synthetic indolicidin analog. Antimicrob Agents Chemother. 1997;41(4):771–775. doi:10.1128/AAC.41.4.771
24. Kazuma K, Ando K, Nihei K-I, et al. Peptidomic analysis of the venom of the solitary bee Xylocopa appendiculata circumvolans. J Venom Anim Toxins Trop Dis. 2017;23:40. doi:10.1186/s40409-017-0130-y
25. Huang HW. DAPTOMYCIN, its membrane-active mechanism vs. that of other antimicrobial peptides. Biochimica et Biophysica Acta (BBA). 2020;1862(10):183395. doi:10.1016/j.bbamem.2020.183395
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.