Back to Journals » Journal of Inflammation Research » Volume 15
The Analysis of the Ubiquitylomic Responses to Streptococcus agalactiae Infection in Bovine Mammary Gland Epithelial Cells
Authors Tong J, Ji X , Zhang H, Xiong B, Cui D, Jiang L
Received 6 April 2022
Accepted for publication 19 July 2022
Published 27 July 2022 Volume 2022:15 Pages 4331—4343
DOI https://doi.org/10.2147/JIR.S368779
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Jinjin Tong,1,* Xintong Ji,1,* Hua Zhang,1 Benhai Xiong,2 Defeng Cui,3 Linshu Jiang1
1Beijing Key Laboratory for Dairy Cow Nutrition, Beijing University of Agriculture, Beijing, People’s Republic of China; 2State Key Laboratory of Animal Nutrition, Institute of Animal Science, Chinese Academy of Agricultural Sciences, Beijing, People’s Republic of China; 3Beijing Key Laboratory of TCVM, Beijing University of Agriculture, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Defeng Cui, Beijing Key Laboratory of TCVM, Beijing University of Agriculture, Beijing, People’s Republic of China, Tel +86 (10)-81798091, Email [email protected] Linshu Jiang, Beijing Key Laboratory for Dairy Cow Nutrition, Beijing University of Agriculture, Beijing, People’s Republic of China, Tel +86 (10)-81798091, Email [email protected]
Purpose: Streptococcus agalactiae is one of the primary pathogens responsible for subclinical mastitis, a significant economic burden for dairy farms. An essential component of the immune response to infection is ubiquitination, which plays important roles in the complex interactions between the pathogen and host.
Materials and Methods: In the present study, quantitative ubiquitylomics was performed to profile changes in the global ubiquitinome of bovine mammary gland epithelial cells (BMECs) infected with S. agalactiae.
Results: The most notable changes in the BMEC ubiquitinome were related to the adherens junction, ribosome, and tight junction pathways. Ubiquitination of CTNNB1, EGFR, ITGB1, CTNNA1, CTNNA2, CDH1, YES1, and SLC9A3R1 appears to be fundamental for regulating multiple cellular processes in BMECs in response to S. agalactiae infection. In addition, broad ubiquitination of various effectors and outer membrane proteins was observed. Ubiquitinated proteins in S. agalactiae-infected BMECs were associated with regulating cell junctions in the host, with potential implications for susceptibility to infection.
Conclusion: The preliminary findings suggest that extensive ubiquitination of CTNNB1, CDH1 and SLC9A3R1 and proteins closely related to cell junctions might play an important role in mastitis progression in dairy cows. The results provide evidence that ubiquitin modification of certain proteins in S. agalactiae-infected BMECs could be a promising therapeutic strategy for reducing mammary gland injury and mastitis.
Keywords: ubiquitination, protein degradation, mastitis, dairy cows
Introduction
Mastitis is characterized by inflammation of the mammary gland. In dairy cows, Streptococcus agalactiae (S. agalactiae) is the predominant causative agent of subclinical mastitis, which negatively affects animal welfare, milk quality, and dairy profitability in addition to increasing the use of antimicrobials.1 Since S. agalactiae is widespread, many strategies have been tested for eliminating the predisposing factors such as environmental modification, feeding, milking equipment, rapid tests for diagnosing subclinical mastitis, antibiograms, treatments during the dry period, etc. A major challenge is that the mechanisms underlying protection against mastitis and its progression have not been fully identified.
Several studies have sought to improve the detection of subclinical mastitis by identifying and quantifying biomarkers that reflect host-pathogen interactions and associated defensive molecular mechanisms. A potential target for biomarker development is ubiquitination, a common posttranslational modification (PTM) that enables rapid proteomic responses to environmental changes.2 Ubiquitination regulates vital cellular functions, including protein degradation, cellular differentiation, transcriptional regulation, and induction of the synthesis of specific proteins.3 Upon bacterial infection, the ubiquitin (Ub) system plays a central role in protecting the nucleus by modifying bacterial effector proteins.4,5 Once nuclear factor-κB (NF-κB) is activated, ubiquitination induces an inflammatory response to inhibit bacterial proliferation. Modification by Ub marks bacteria for elimination by proteasome-, phagolysosome-, and autophagosome-regulated degradation pathways.6 Ubiquitylation also regulates numerous cellular functions, including protein-protein interactions, signal transduction, receptor internalization, and subcellular localization of biomarker proteins.7 A recent study revealed that in infected epithelial cells, a small number of Salmonella escape the vacuoles and enter the cytosol, labeled with a thick Ub coat.8 However, the proteins responsible for recognizing and ubiquitinating bacteria during mastitis have not been fully identified, and the ubiquitination sites on the surface of S. agalactiae remain unclear.
Improvements in mass spectrometry (MS)-based proteomics approaches for characterizing endogenous ubiquitinated peptides9–11 have facilitated protein ubiquitination detection and quantification. Nonetheless, efforts to characterize protein changes in milk, mammary gland epithelial cells, and tissues during clinical or subclinical mastitis have yielded inconclusive results, and no biomarker or method for detecting subclinical mastitis in the early stages with high accuracy and precision is available. While ubiquitylomics has been utilized effectively to evaluate stimulus-dependent changes in the cellular ubiquitinome6 comprehensive information on the cell ubiquitinome upon S. agalactiae infection is lacking.
To address these gaps, we utilized ubiquitylomics to systematically and quantitatively assess the changes in the ubiquitinome of S. agalactiae-infected bovine mammary gland epithelial cells (BMECs). The results represent an unparalleled resource of Ub-modified targets and changes in S. agalactiae-infected BMECs that can be used in future studies of milk synthesis, mammary biology, and immune regulation in the mammary gland and to establish new biomarkers for the early diagnosis of subclinical mastitis.
Materials and Methods
Cell Culture and S. agalactiae Infection
Primary BMECs were cultured as previously described12 in Dulbecco’s modified Eagle’s medium/F-12 (DMEM/F-12, Gibco) supplemented with 15% fetal bovine serum (FBS, Gibco), 100 U/mL penicillin, and 100 μg/ mL streptomycin at 37 °C in a humidified atmosphere of 5% CO2. The Beijing University of Agriculture Animal Care and Use Committee (BUA2021017, Beijing, China) approved the experimental procedures. All the bovine mammary epithelial cells were used in the present study after three passages. Cells were grown in 6-well plates (1x106 cells/well) to ~80% confluence and then infected with S. agalactiae (CVCC 3940) at a multiplicity of infection (MOI) of 50 for 6 h to characterize the ubiquitinome of S. agalactiae-infected BMECs. Cells without S. agalactiae infection were used as the control group. Each group had three biological replicates. As reported in our previous study, the cells were harvested at specific time points in denaturing lysis buffer and pooled in equivalent proportions depending on protein concentration.13
Protein Extraction and Digestion
BMECs were sonicated three times on ice using a high-intensity ultrasonic processor (Scientz) in lysis buffer (8 M urea, 1% protease inhibitor cocktail, 50 μM PR-619). After removing debris by centrifugation at 12,000 g and 4 °C for 10 min, the protein concentration in the supernatant was analyzed with a BCA kit (Beyotime) following the manufacturer’s instructions. Equal quantities of each sample were enzymatically hydrolyzed, and the volume was adjusted to be consistent with the lysate. Pre-cooled trichloroacetic acid (TCA) was gradually added to the samples to a final concentration of 20% to precipitate protein, followed by vortexing to mix and incubation for 2 h at 4°C. The precipitates were collected by centrifugation at 4500 g for 5 min at 4°C. The precipitated proteins were washed three times with cold acetone by centrifugation for 5 min at 4500 g and 4°C. After drying the precipitate, TEAB was added at a final concentration of 200 mM, and sonication was performed multiple times on ice by a high-intensity ultrasonic processor (Scientz, Ningbo, China). Next, trypsin was added at a trypsin/protein mass ratio of 1:50 and incubated overnight. Disulfide bonds were reduced by incubation with 5 mM DTT for 30 min at 56°C, followed by alkylation with 11 mM indole-3-acetic acid (IAA) for 15 min at room temperature protected from light. Finally, the peptides were desalted with a Strata C18 SPE column (Phenomenex), and the peptide concentration was measured with a NanoPhotometer (IMPLEN). Equal quantities of each peptide sample were dried by vacuum centrifugation; 2 mg of the peptide was required for ubiquitination analysis.
Lysine Ubiquitylated Peptide Enrichment
The tryptic peptides were dissolved in NTEN buffer (100 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 0.5% NP-40, pH 8.0) and incubated with prewashed anti-K-e-GG beads (PTM-1104; PTM Biolabs, Hang-zhou, China) at 4°C overnight with moderate shaking. After washing four times with NTEN buffer and twice with ddH2O, the beads were eluted with 0.1% (v/v) trifluoroacetic acid, and the eluates were vacuum-dried. The peptides were then desalted with C18 ZipTips (Millipore, Billerica, MA), following the manufacturer’s instructions.
LC-MS/MS Analysis
The liquid chromatography-tandem mass spectrometry (LC-MS/MS) equipment comprised a NanoElute UHPLC system linked to a hybrid trapped ion mobility spectrometry–quadrupole time of flight mass spectrometer (timsTOF Pro, Bruker Daltonics, Bremen, Germany) via a nanospray electron ionization source. The C18 reversed-phase column (Thermo Scientific Easy Column) was equilibrated with buffer A (2% acetonitrile and 0.1% formic acid) and loaded with peptide sample in buffer A via an autosampler. Buffer B was 100% acetonitrile and 0.1% formic acid, and the flow rate was 300 nl/min. Peptides were eluted with a linear gradient from 7 to 24% B in 42 min, followed by an increase to 32% B in 12 min and a subsequent increase to 80% in 3 min; 80% B was then maintained for 3 min. The fraction of buffer B was increased to 95% within 1 min, followed by washing and column equilibration with buffer A for 15 min to regenerate the column for the next sample. The peptides were isolated by the ultra-performance liquid phase system and subsequently infused into the capillary ion source for ionization and analysis by timsTOF Pro mass spectrometry. The peptides were ionized at an applied voltage of 1.6 kV. The peptide parent ions and their secondary fragments were detected and analyzed using high-resolution TOF. The MS information was obtained using a data-dependent top 10 method that dynamically chose the most abundant precursor ions from the survey scan (400–1500 m/z) for Online Parallel Accumulation–Serial Fragmentation (PASEF) mode. 10 PASEF were gathered after each full scan to collect the mass-to-charge ratios of the peptides and peptide fragments, and the parent ion was identified by a charge number in the 0–5 range by secondary mass spectrometry. The target value was determined based on predictive Automatic Gain Control. The dynamic exclusion time was 30s.
Ubiquitination Motif Analysis
Potential ubiquitination motifs were identified as described previously by Zhu et al by applying Motif-x14 to the ubiquitinome data of S. agalactiae-infected BMECs. 20-mer peptides comprising the ten upstream and downstream residues flanking the ubiquitination sites were acquired for internal ubiquitination sites. If the protein ubiquitination site was at the N- or C-terminus, the ubiquitinated peptide was supplemented with 10-mers with the required number of “x” rather than any amino acid. The parameters were “pre-aligned”, central K, width = 21, occurrences = 20, and significance = 0.000001, and the protein sequences of Bos taurus in UniProt were uploaded as the background.
Sequence Database Searching and Data Analysis
MaxQuant (version 1.6.15.0) was used to analyze the MS data. The MS data were searched against 37,512 sequences from Uniprot_Bos_taurus_9913_PR_20201217.fasta to identify proteins and ubiquitination sites from tandem mass spectra; reverse decoy, and common contaminant protein sequences were utilized as false-positive controls. Trypsin/P was the cleavage enzyme, and up to 4 missing cleavages were permitted. The minimum peptide length was set at 7, and 5 modifications per peptide were permitted. The mass error was set to 20 ppm for precursor ions, and the fragment ion tolerance was set to 0.02 Da (Supplementary Figure 1, 2 and 3A-B). Carbamidomethylation (C) was set as a fixed modification, while oxidation on methionine and ubiquitination on lysine and N-terminal methionine were variable modifications. The false discovery rate (FDR) threshold for identifying proteins, peptides, and modification sites was set to 1%. All other parameters were set to their default values in MaxQuant. Potential Kub sites with a localization probability of less than 75% and reverse or contaminant sequences were omitted.
Western Blot
Western blot analyses were performed as previously described (35). Briefly, cells were washed 3 times with PBS and lysed at 4°C in RIPA buffer (P0013C, Beyotime Biotechnology) containing phenylmethylsulfonyl fluoride (0.5 mM, ST506, Beyotime Biotechnology), aprotinin (5 μg/mL, A1153, Sigma-Aldrich), and leupeptin (5 μg/mL, L2884, Sigma-Aldrich). Approximately 50 μg of protein was separated by 8% SDS-PAGE and blotted onto a nitrocellulose membrane. The membrane was blocked with 5% skim milk in Tris-buffered saline with 0.05% Tween 20 for 1 h at room temperature and then incubated overnight at 4°C with primary antibodies at the following dilutions: p65 (1:1000 dilution; 6956), p38 (1:1000 dilution; 14,451), JNK (1:1000 dilution; 9252), p-p38 (1:1000 dilution; 9215s), p-JNK (1:1000 dilution; 9251), p-ERK1/2 (1:1000 dilution; 4370T) and β-actin (1:1000 dilution; 4970, Cell Signaling Technology). The blots were then incubated for 1 h at room temperature with the appropriate secondary antibodies conjugated to horseradish peroxidase (1:2000 dilution; ZSGB-Bio). Proteins were visualized using an enhanced chemiluminescent detection reagent (6683, Signaling Technology). Densitometric analysis was performed with Image-J software, and target protein expression was normalized to β-actin expression.
Statistic and Bioinformatics Analysis
Results are presented as the mean ± standard error of the mean of triplicate samples in each experimental group. Statistical analyses were performed in GraphPad Prism 6 (GraphPad Software Inc.). For Western blots, the statistical significance of differences was assessed using Student’s t-test. Differences were considered significant at P < 0.05.
Online tools (http://bioinformatics.psb.ugent.be/webtools/Venn/) were used to create a Venn diagram plotting the overlap of proteins and peptides among the three biological replicates. Comparisons were performed using the relative ubiquitination abundance, which was determined by dividing the intensity of an individual peptide by the mean intensity. Only ubiquitinated peptides with consistent fold changes in at least two of the three replicates were considered. Proteins or peptides were considered differentially expressed when the difference in abundance among samples was > 1.5-fold. Wilcoxon analysis was used for comparisons, and P < 0.05 was considered statistically significant. P values were corrected for the FDR in each dataset. Protein pathways were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.kegg.jp/) database. InterPro (http://www.ebi.ac.uk/interpro/) was utilized for protein domain annotation. WoLF PSORT (PSORT/PSORT II, http://www.genscript.com/wolf-psort.html) was used for subcellular localization prediction. The protein sequences in the Bos taurus database served as the background database parameters. Secondary structure types were determined for Kub (ubiquitinated lysine) positions using NETSURFP (version 1.1). For category enrichment, Fisher’s exact test was utilized with P < 0.05. Functional interaction network analysis was performed using STRING software and visualized by Cytoscape (version 3.4.0). Bioedit (version 7.2.5) was utilized for sequence alignments with Bos taurus.
Results and Discussion
Large-scale profiling of ubiquitination sites in S. agalactiae-infected BMECs
To comprehensively identify lysine ubiquitinated (Kub) sites in BMECs infected with S. agalactiae, we extracted proteins from BMECs infected with S. agalactiae (MOI 50:1, 6 h) and from cells that were not infected as described in our previous study.12 Protein ubiquitination was quantified by combining label-free immunoaffinity enrichment with high-resolution mass spectrometry and a high-quality anti-K-ε-GG antibody (PTM Biolabs). Briefly, after trypsin digestion, Kub-containing peptides were enriched by affinity purification with Kub antibodies and analyzed by LC-MS/MS regarding the UniProt database (Figure 1A). Significantly different (P < 0.05 by the Student’s t-test) proteins were characterized, and proteins and ubiquitination sites with a fold change of 1.5 or greater were considered differentially regulated by S. agalactiae infection. In total, 1805 up-regulated sites, 879 up-regulated proteins, 904 down-regulated sites, and 484 down-regulated proteins were identified (Figure 1B). Overall, we identified 4121 proteins and 18,443 sites in the three biological replicates, of which 2686 proteins and 9108 sites were quantified (Figure 1C).
Quantitative Proteomics of the Ub-Modified Proteome and Analysis
Among the detected ubiquitinated proteins, a total of 18,443 Kub peptides from 4121 unique proteins with peptide scores > 40 were identified. The peptide length distribution of the ubiquitinated peptides was not uniform (Figure 2A). The percentages of identified ubiquitinated peptides with 1, 2, 3, and 4 ubiquitination sites were 78.50% (14,478/18,443), 20.32% (3748/18,843), 1.10% (202/18,843) and 0.08% (15/18,843), respectively. Most of the identified proteins had a single ubiquitination site (34.41%, 1418/4121) (Figure 2B), but some proteins in S. agalactiae-infected BMECs had more than 20 ubiquitination sites (2.27%, 114/4121) (Figure 2C). These results are consistent with previous studies using similar methodologies,15–17 confirming that the mass spectrometry quantification platform and strategy were reliable and efficient.
Next, we further examined the relationship between the ubiquitinome and proteome. Among the identified ubiquitylated proteins, 1823 were also present in the proteomics dataset (Figure 2D). Among the up-regulated ubiquitinated proteins, 33 were down-regulated in the proteome (Figure 2E), whereas only Q58DK4 was down-regulated in the ubiquitinome but up-regulated in the proteome. RPL7 and RPL7A overlapped among the up-regulated proteins, up-regulated ubiquitinated proteins, and down-regulated ubiquitinated proteins, and 10 proteins overlapped among the down-regulated proteins, up-regulated ubiquitinated proteins, and down-regulated ubiquitinated proteins. RPL7 is an essential 60S preribosomal assembly factor and plays an important role in inhibiting ribosome translation.18 These sets of ubiquitinated proteins will provide a foundation for unraveling the posttranslational regulation of the response of BMECs to S. agalactiae infection.
Sequence Properties of Ubiquitinated Proteins
To further characterize the amino acid sequences surrounding ubiquitination sites in BMECs infected with S. agalactiae, the position-specific frequencies of amino acid residues neighboring Kub sites were analyzed using the program MOMO. From the set of 18,338 Kub peptides, we identified five conserved motifs among 5876 unique sites, which represented approximately 32% of the identified sites (Supplementary Table S1). In these five motifs, two residues were highly enriched upstream or downstream of Kub sites (Figure 3A): glutamic acid (E) and leucine (L) (Figure 3A). We also observed a significant preference for hydrophobic residues like leucine, which are important in mammary gland epithelial cells.19 The five motifs were xxxxxxxxxE_K_xxxxxxxxxx, xxxxxxxxxx_K_xLExxxxxxx, xxxxxxxxxx_K_Exxxxxxxxx, xxxxxxxxxx_K_LxxxxxxRxx, and xxxxxxxxxx_K_xxExxxxxxx (x indicates any amino acid) and varied in abundance (Figure 3B). The motif xxxxxxxxxE_K_xxxxxxxxxx was described previously20 in young rice leaves. The other four motifs are novel and thus may provide insights on the mechanism of mastitis, particularly in response to S. agalactiae infection.
To evaluate the relative abundances of different amino acids surrounding Kub sites,21 the parameters “pre-aligned”, central K, width = 21, occurrences = 20, and significance=0.000001 were applied using protein sequences from cattle (Bos_taurus_9913_PR_20201217.fasta) in UniProt as a reference (Figure 3C). This analysis revealed a significant preference for hydrophilic residues such as Glu(E) at positions adjacent to Kub sites. In addition, E was the residue with the greatest frequency in position –1 and was also enriched at positions +1 and +3. Thus, the sequence preferences in the heatmap data were consistent with the motif analysis. Moreover, the observed preference for hydrophobic residues such as Leu(L) adjacent to Kub sites agrees with a previous analysis of mammalian sequences.22 In mastitis milk samples, the percentages of lysine, leucine, and alanine increase.23 Notably, in response to leucine, leucyl-tRNA synthetase initiates the mTORC1 pathway by acting as a GTPase-activating protein for a component of mTORC1, thereby transducing amino acid availability to mTOR signaling.24
Consequently, it can be speculated that leucine is also closely related to milk protein synthesis in the mammary gland through mTOR signaling. By contrast, the basophilic residue arginine (R) was excluded from neighboring positions. Overall, these results reveal differences in Kub motifs in S. agalactiae-infected BMECs compared with non-infected cells.
In addition to the primary sequence, the secondary structure is important for Kub site prediction.25,26 We, therefore, used the software NetSurfP to incorporate protein secondary structural features. The probabilities of various secondary structures (curl, α-helix, and β-strand) near Kub sites were compared with the secondary structure probabilities for all lysine sites in identified proteins. Kub sites were preferentially located in α-helices over β-strands (p=9.63E-14 for α-helix and p=2.59E-36 for β-strand)(Figure 4). In addition, the secondary structures of both ubiquitinated and non-ubiquitinated sites were enriched in coiled coils, with an average of approximately 60%. These results imply that lysines are selectively ubiquitinated to alter protein structure and activity, which may influence the functions of the modified proteins in S. agalactiae-infected BMECs.
 |
Figure 4 Distribution of secondary structures of ubiquitination and non-ubiquitination sites: α-helix, β-strand, and coil. Significance was calculated by the Wilcoxon test. |
Annotation of the S. agalactiae-Regulated Ubiquitinome in BMECs
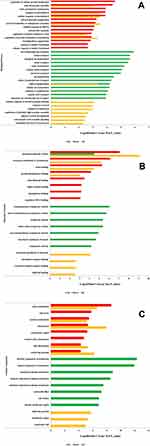
To further elucidate the biological functions of ubiquitination in BMECs infected with S. agalactiae, Gene Ontology (GO) enrichment analysis of the identified ubiquitinated proteins was performed. Analysis of the biological process category revealed that the up-regulated ubiquitinated proteins were highly enriched in regulation of cellular protein localization, cell-cell junction assembly, and actin cytoskeleton organization, whereas the down-regulated ubiquitinated proteins were enriched in ion transmembrane transport, cation transport, and inorganic ion homeostasis (Figure 5A and Table S2). Actin cytoskeleton organization and cell-cell adhesion influence the cell adhesion properties of cells, and changes in cell adhesion have been directly or indirectly associated with lactation and immune and inflammatory responses.27–29 The results of the GO enrichment analysis suggest that S. agalactiae infection of BMECs damages cell junctions and disrupts pathways related to immune function and lactation.
To further assess whether cell junction- and inflammation-related proteins involved in S. agalactiae infection induce the transcription of related cell adhesion pathways in BMECs, we performed Western blotting (Supplementary Figure 4). Compared with the control, S. agalactiae infection significantly decreased the protein levels of p65, JNK, p-JNK, p-ERK1/2, P38, and p-P38. It has been reported that changes in ROS-p38 signaling in Drosophila pericardial cells alter the abundance of septate junction proteins and the proteins Coracle and Kune-Kune.30 Furthermore, p38 activation plays an important role in regulating the flagellin-induced innate immune response in airway epithelia during infection with the human pathogen Pseudomonas aeruginosa.31 Interestingly, in line with our Western blot results, Li32 recently found that deoxynivalenol accelerates the endocytosis and degradation of tight junction proteins and that this process is regulated by the activation of p38 MAPK signaling pathways by a c-Jun-N-terminal kinase (JNK). Lithium enhances post-stroke blood-brain barrier integrity by regulating tight junctions and has also been identified as a key regulator of the prosurvival MAPK/ERK1/2 pathway.33 Altogether, these findings suggest that cell junction proteins may have important roles beyond their canonical barrier function, especially those ubiquitinated in BMECs in response to S. agalactiae infection.
In the category of molecular functions, the up-regulated ubiquitinated proteins were enriched in structural molecule activity, structural constituent of the cytoskeleton, and actin-binding, whereas the down-regulated ubiquitinated proteins were enriched in transmembrane transporter activity, dynamic transmembrane transporter activity, and symporter activity (Figure 5B). In the cellular component category, the up-regulated ubiquitinated proteins were highly enriched in regulation of actin cytoskeleton, cell cortex, cortical cytoskeleton, and cell junction, and the down-regulated ubiquitinated proteins were enriched in an intrinsic component of membrane, an integral component of the membrane and basolateral plasma membrane (Figure 5C). Dysregulation of ubiquitinated proteins has been reported to impact cell development and proliferation. Cell movement, a type of cell migration, is closely related to inflammation.28,34 Collectively, these findings indicate that ubiquitinated proteins in S. agalactiae-infected cells are closely related to cell adhesion, cell junctions, regulation of biological processes, and maintaining the balance between apoptosis and granulopoiesis in cell turnover, suggesting potential mechanisms underlying susceptibility or resistance to intramammary infection.
Functional Enrichment Analysis of the Identified Ubiquitinated Proteins
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed to clarify the functions of the ubiquitinated proteins (Figure 6A). The most significantly enriched pathways were adherens junction (P = 4.08488E-05), ribosome (P = 4.08488E-05), and tight junction (P = 4.08488E-05), in addition to other biosynthesis and metabolism pathways. Cell adhesion-related pathways are involved in mammary tissue morphology, secretion composition, lactation, and tight junction integrity.35,36 The complex mammary response to bacterial infection is characterized by changes in morphology and immune regulation in epithelial cells and mammary tissue, changes in the composition of mammary secretions, and changes in the integrity of tight junctions.37,38 The enrichment of pathways related to cell junction regulation in S. agalactiae-infected BMECs is consistent with these observations.
Because protein domains are conserved protein sequences that frequently represent structural units, we next performed statistical analysis to identify domains enriched in the ubiquitinome of S. agalactiae-infected BMECs. Twenty-three protein domains were significantly enhanced (P < 0.05). The top 10 domains were FERM N-terminal domain, FERM central domain, FERM C-terminal PH-like domain, UBA/TS-N domain, proteasome subunit A N-terminal signature, spectrin repeat, PDZ domain (also known as DHR or GLGF), cadherin domain, myosin head (motor domain) and EF-hand domain pair (Figure 6B). These results suggest that the ubiquitinome includes proteins involved in numerous fundamental cellular processes, like protein degradation and secondary metabolism.
Using the STRING database, we eliminated all background proteins in all pathways regulated by ubiquitination to more clearly visualize the PPIs (protein-protein interactions) between the significantly altered proteins (Figure 7A). Next, the top 5 KEGG pathways were identified for further examination (Figure 7B). PPI network analysis revealed a wide range of interactions influenced by protein ubiquitination. In particular, ubiquitination of CTNNB1, EGFR, ITGB1, CTNNA1, CTNNA2, CDH1, YES1, and SLC9A3R1 appears to play an essential role in regulating numerous cellular processes in BMECs infected with S. agalactiae.
Our previous study showed that CTNNB1(β-catenin) and CDH1(E-cadherin) play an important role in the Wnt signaling pathway by activating CyclinD1 and cell proliferation in bovine mammary epithelial cells.39 Furthermore, growing evidence has shown that ITGB1 (β1-integrin) regulates mammary gland development and function, closely related to prolactin signaling.40,41,42 Importantly, the SLC9A3R1-related signaling pathway regulates the activation of autophagy processes 43. Further research on mammary gland mastitis is warranted. Several lines of evidence indicate that various membrane proteins in BMECs, including immune sensors (Toll-like receptors, TLR), nutrient transporters (glucose transporter and aquaporin), and tight junction proteins (claudin and occludin), are involved in the onset of mastitis or milk production.39,40
Previous studies applying omics technologies to dairy cows have largely omitted posttranslational regulation.41,42 The present extensive proteomic investigation of the ubiquitinome in BMECs infected with S. agalactiae provides insights on the regulatory roles and mechanisms of protein ubiquitination in the mammary gland in dairy cows and a reference for the development of novel biomarkers for mastitis therapy.
Data Sharing Statement
There is no limitation on data availability. The ubiquitin and global proteomics data can be downloaded and viewed using the following Shiny Web applications: https://vilchezlab.shinyapps.io/shiny-volcanoplot/ and https://vilchezlab.shinyapps.io/shiny-heatmap/. Proteomics information has been deposited in the Proteome Xchange Consortium through the PRIDE partner repository with the dataset identifiers PXD028659 (ubiquitin proteomics of BMECs infected or not infected with S. agalactiae) and PXD028585 (global protein proteomics of BMECs infected or not infected with S. agalactiae).
Ethics Approval and Informed Consent
The Beijing University of Agriculture Animal Care and Use Committee (BUA2021017, Beijing, China) approved all of the experimental procedures.
Acknowledgments
This study was financially supported by the Project of R&D Program of Beijing Municipal Education Commission(KM202210020006) and the Key Project of Beijing Municipal Education Commission (20JF0008).
Disclosure
The authors declared no conflicts of interest regarding this work.
References
1. Pascal R, Gilbert FB, Pierre G, et al. Invited review: a critical appraisal of mastitis vaccines for dairy cows. J Dairy Sci. 2021;104:10427–10448.
2. Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nature Structural Mol Biol. 2011;18:520–528.
3. Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12:295–307.
4. Tanner K, Brzovic P, Rohde JR. The bacterial pathogen-ubiquitin interface: lessons learned from Shigella. Cell Microbiol. 2014;17:35–44.
5. Ashida H, Kim M, Sasakawa C. Exploitation of the host ubiquitin system by human bacterial pathogens. Nat Rev Microbiol. 2014;12:399.
6. Fiskin E, Bionda T, Dikic I, et al. Global Analysis of Host and Bacterial Ubiquitinome in Response to Salmonella Typhimurium Infection. Mol Cell. 2016;62:967–981.
7. Majolée J, Kovaevi I, Hordijk PL. Ubiquitin-based modifications in endothelial cell–cell contact and inflammation. J Cell Sci. 2019;132:854.
8. Huett A, Heath RJ, Begun J, et al. The LRR and RING Domain Protein LRSAM1 Is an E3 Ligase Crucial for Ubiquitin-Dependent Autophagy of Intracellular Salmonella Typhimurium. Cell Host Microbe. 2012;12:778–790.
9. Ordureau A, Paulo JA, Zhang J, et al. Global Landscape and Dynamics of Parkin and USP30-Dependent Ubiquitylomes in iNeurons during Mitophagic Signaling. Elsevier Sponsored Documents. 2020;1;77.
10. Li X, Zhang C, Zhao T, et al. Lysine-222 succinylation reduces lysosomal degradation of lactate dehydrogenase a and is increased in gastric cancer. J Exp Clin Cancer Res. 2020;39:172.
11. Wang Y, Cheng X, Yang T, et al. Nitrogen-Regulated Theanine and Flavonoid Biosynthesis in Tea Plant Roots: protein-Level Regulation Revealed by Multiomics Analyses. J Agr Food Chem. 2021;69:10002–10016.
12. Tong J, Li Y, Liu R, et al. Effect of Semen vaccariae and Taraxacu mogono on Cell Adhesion of Bovine Mammary Epithelial Cells. J Integrative Agriculture. 2012;11:2043–2050.
13. Tong J, Sun M, Zhang H, et al. Proteomic analysis of bovine mammary epithelial cells after in vitro incubation with S. agalactiae: potential biomarkers. Vet Res. 2020;51:98.
14. Chou MF, Schwartz D. Biological sequence motif discovery using motif-x. Curr Protocols Bioinformatics. 2011;35:1–24.
15. Li W, Wang H, Yang Y, et al. Integrative Analysis of Proteome and Ubiquitylome Reveals Unique Features of Lysosomal and Endocytic Pathways in Gefitinib‐Resistant Non‐Small Cell Lung Cancer Cells. PROTEOMICS. 2018;6:e1700388.
16. Dybas JM, O’Leary CE, Ding H, et al. Integrative proteomics reveals an increase in non-degradative ubiquitylation in activated CD4 T cells. Nat Immunol. 2019;20:1–9.
17. Sun Y, Zheng X, Yuan H, et al. Proteomic analyses reveal divergent ubiquitylation patterns in hepatocellula carcinoma cell lines with different metastasis potential. J Proteomics. 2020;225:103834.
18. Tang Z. Sp1-Mediated circRNA circHipk2 Regulates Myogenesis by Targeting Ribosomal Protein Rpl7. Genes. 2021;12:696–708.
19. Kishi M, Itagaki Y, Sudo T, et al. In vitro development of bovine nuclear transfer embryos reconstructed with mammary gland epithelial cells at different passages. Animal Sci J. 2003;74:363–368.
20. Xie X, Kang HX, Liu WD, et al. Comprehensive Profiling of the Rice Ubiquitome Reveals the Significance of Lysine Ubiquitination in Young Leaves. J Proteome Res. 2015;14:2017–2025.
21. Deng W, Wang Y, Liu Z, et al. HemI: a Toolkit for Illustrating Heatmaps. PLoS One. 2014;9:e111988.
22. Wagner SA, Beli P, Weinert BT, et al. A Proteome-wide, Quantitative Survey of In Vivo Ubiquitylation Sites Reveals Widespread Regulatory Roles. Mol Cell Proteomics Mcp. 2011;10:
23. Andrei S, Culea M, Matei S, et al. Amino Acid Concentration in Normal and Subclinical Mastitis Milk. Physics of Fluids. 2011;23:2154.
24. Han J, Jeong S, Park M, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424.
25. Muller J, Szklarczyk D, Julien P, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2010;38:D190–D195.
26. Florian G, Jeremy G, Matthias M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 2011;39:253–260.
27. Banos G, Bramis G, Bush SJ, et al. The genomic architecture of mastitis resistance in dairy sheep. Bmc Genomics. 2017;18:624.
28. Lopreiato V, Minuti A, Morittu VM, et al. Short communication: inflammation, migration, and cell-cell interaction-related gene network expression in leukocytes is enhanced in Simmental compared with Holstein dairy cows after calving. J Dairy Sci. 2020;103:1908–1913.
29. Richards VP, Choi SC, Pavinski Bitar PD, et al. Transcriptomic and genomic evidence for Streptococcus agalactiae adaptation to the bovine environment. BMC Genomics. 2013;14:920–932.
30. Lim HY, Bao H, Liu Y, et al. Select Septate Junction Proteins Direct ROS-Mediated Paracrine Regulation of Drosophila Cardiac Function. Cell Rep. 2019;28:1455–1470.e1454.
31. Illek B, Fu Z, Schwarzer C, et al. Flagellin-stimulated Cl- secretion and innate immune responses in airway epithelia: role for p38. Am j Physiol Lung Cell Mol Physiol. 2008;295:531–542.
32. Li E, Horn N, Ajuwon KM. Mechanisms of deoxynivalenol-induced endocytosis and degradation of tight junction proteins in jejunal IPEC-J2 cells involve selective activation of the MAPK pathways. Arch Toxicol. 2021;95:2065–2079.
33. Mh A, Bz A, Bbbc D, et al. Lithium enhances post-stroke blood-brain barrier integrity, activates the MAPK/ERK1/2 pathway and alters immune cell migration in mice - ScienceDirect. Neuropharmacology. 2020;181:1–14.
34. Asselstine V, Miglior F, Suárez-Vega A, et al. Genetic mechanisms regulating the host response during mastitis. J Dairy Sci. 2019;102:9043–9059.
35. Huang J, Luo G, Zhang Z, et al. iTRAQ-proteomics and bioinformatics analyses of mammary tissue from cows with clinical mastitis due to natural infection with Staphylococci aureus. BMC Genomics. 2014;15:1–14.
36. Zhao X, Ponchon B, Lanctt S, et al. Invited review: accelerating mammary gland involution after drying-off in dairy cattle. J Dairy Sci. 2019;102:6701–6717.
37. Oviedo-Boyso J, Valdez-Alarcón JJ, Cajero-Juárez M, et al. Innate immune response of bovine mammary gland to pathogenic bacteria responsible for mastitis. J Infect. 2007;54:399–409.
38. Swartz TH, Bradford BJ, Mamedova LK. Diverging in vitro inflammatory responses toward Streptococcus uberis in mouse macrophages either preconditioned or continuously treated with β-hydroxybutyrate. JDS Communications. 2021;1:84.
39. Stumpf MT, Fischer V, Daltro DS, et al. Mammary gland cell’s tight junction permeability from dairy cows producing stable or unstable milk in the ethanol test. Int J Biometeorol. 2020;64:1981–1983.
40. Tsugami Y, Wakasa H, Kawahara M, et al. Adverse effects of LPS on membrane proteins in lactating bovine mammary epithelial cells. Cell Tissue Res. 2021;384:435–448.
41. Mudaliar M, Tassi R, Thomas F, et al. Mastitomics, the integrated omics of bovine milk in an experimental model of Streptococcus uberis mastitis: 2. Label-free relative quantitative proteomics. Mol Biosyst. 2016;12:2748–2761.
42. Mudaliar M, Thomas FC, Eckersall PD. Omic Approaches to a Better Understanding of Mastitis in Dairy Cows. Periparturient Diseases Dairy Cows. 2017;12:139–183.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.