Back to Journals » Cancer Management and Research » Volume 12
The Analysis of the Anti-Tumor Mechanism of Ursolic Acid Using Connectively Map Approach in Breast Cancer Cells Line MCF-7
Authors Guo W, Xu B, Wang X, Zheng B, Du J, Liu S
Received 11 December 2019
Accepted for publication 23 April 2020
Published 15 May 2020 Volume 2020:12 Pages 3469—3476
DOI https://doi.org/10.2147/CMAR.S241957
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Weiqiang Guo,1 Bin Xu,1 Xiaoxiao Wang,2 Bo Zheng,1 Jiahui Du,2 Songbai Liu2
1School of Chemistry, Biology and Material Engineering, Suzhou University of Science and Technology, Suzhou 215009, People’s Republic of China; 2Suzhou Key Laboratory for Medical Biotechnology, Suzhou Vocational Health College, Suzhou 215009, People’s Republic of China
Correspondence: Weiqiang Guo
School of Chemistry, Biology and Material Engineering, Suzhou University of Science and Technology, Suzhou 215009, People’s Republic of China
Email [email protected]
Songbai Liu
Suzhou Key Laboratory for Medical Biotechnology, Suzhou Vocational Health College, Suzhou 215009, People’s Republic of China
Email [email protected]
Background: Ursolic acid (UA), a primary bioactive triterpenoid, was reported as an anti-cancer agent. However, the current knowledge of UA and its potential anti-cancer mechanisms and targets in breast cancer cells are limited. In this study, we aimed to illustrate the potential mechanisms and targets of UA in breast cancer cells MCF-7.
Methods: The effect of UA on cell growth was determined in MCF-7 cells by MTT assay. The anti-tumor mechanism of UA was evaluated by microarray, CAMP, and Western blot. Moreover, the molecular docking between UA and potential receptors were predicted by iGEMDOCK software.
Results: The result of MTT assay demonstrated that UA could inhibit MCF-7 cell growth with IC50 values of 20 μM. Microarray and CMAP analysis, validated by Western blot, indicated that UA significantly modulated IKK/NF-κB, RAF/ERK pathways, and down-regulated the phosphorylation level of PLK1 in MCF-7 cells.
Conclusion: Our data indicated that the anti-tumor effects of UA are due to the inhibited RAF/ERK pathway and IKK/NF-κB pathway. It could also be explained by the reduced phosphorylation of PLK1 in MCF-7 cells. This study provides a new insight for deep understanding of the new anti-cancer mechanisms of UA in MCF-7 breast cancer cells.
Keywords: ursolic acid, connectivity map, RAF/ERK, IKK/NF-κB, PLK1
Introduction
Breast cancer (BC) is a severe health problem worldwide and ranked as the second cause of cancer-associated deaths in females.1 According to the American cancer society statistics report in 2018, there were 26612 new cases and 40920 deaths of BC.2 In general, BC is classified into different types by the expression level of estrogen receptor (ER), epidermal growth factor receptor (HER2), and progesterone receptor (PR). Different types of BC respond in different ways to various types of treatment, making it more challenging to develop accurate therapeutic methods and drugs that are both effective and safe to BC patients.3,4 Hence, huge efforts are required for developing more effective therapeutic agents.
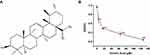
A lot of natural products have been reported to have anti-cancer effects in various types of cancer.5,6 Ursolic acid (UA) (Figure 1A) is a pentacyclic triterpenoid compound extracted from naturally growing herbs, such as apples, pears, and loquat.7,8 Several studies have confirmed that UA has shown strong anticancer effects against lung cancer, BC, and so on.9,10 Yeh et al investigated that UA inhibited cancer cell invasion, migration, and proliferation in MDA-MB-231 via down-regulating u-PA and MMP-2 expression, and up-regulating the expression of PAI-I and TIMP-2.11 UA has also been reported to reduce the phosphorylation of AKT/mTOR signaling for modulating proliferation and apoptosis.12,13 Shishodia et al found that UA had the potential to regulate the cytokines signaling cascade by inhibiting NF-κB stimulation.14 While these studies provided a foundation of possible mechanisms of UA, its precise molecular anti-BC mechanisms and target molecules remain unclear.
Recently, connectivity map (CMAP) database has frequently been employed to resolve the molecular mechanisms and target prediction of drugs.15,16 Liu et al predicted novel histone deacetylase inhibitors by CMAP.17 Lee et al elucidated the potential molecular mechanisms of berberine through CMAP approach.18 Thus, CMAP-based strategies can dramatically streamline the molecular mechanisms and target molecules prediction process for drugs or potential active ingredients. The current study investigated the precise molecular mechanism of UA using CMAP approach. The results suggested that UA could inhibit PLK1, IKK/NF-κB, and RAF/ERK pathways in MCF-7. Our finding provided an extensive view of the anti-tumor mechanisms of UA.
In the present study, the potential molecular mechanisms of UA were analyzed by measuring the gene expression profiles of MCF-7 cells incubated by UA using CMAP approach. Then, the results of CMAP analysis were further verified (validated) by Western blot. Our results demonstrated that UA may inhibit PLK1, IKK/NF-κB, and RAF/ERK pathways. Our finding provided an extensive view of the anti-tumor mechanisms of UA.
Materials and Methods
Materials
UA was purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). Human breast cancer MCF-7 cells were obtained from Shanghai Cell Bank of Chinese Academy of Science (Shanghai, China). Trizol was from Invitrogen (CA, USA). Microarray analysis was finished by Oebiotech (Shanghai, China). Specific antibodies for GAPDH, BRAF, PLK1, IKKβ, NF-κB p65, ERK1/2, p-ERK1/2, p-IKKβ, p-BRAF, p-NF-κB p65, and p-PLK1 were purchased from Abcam (Cambridge, UK).
Cell Culture
MCF-7 cells were maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100U/mL penicillin and 100mg/mL streptomycin at 37°C with a humidified atmosphere of 5% CO2. DMEM, FBS, and antibiotics were purchased from Gibco, Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
Cell Viability Assay
MCF-7 cells were inoculated in 96-well plates (104/well). When cells were in the logarithmic growth period, different concentrations of UA (from 0 to 64 μM) were added. After the cells were treated for 24 h, 20 μL of MTT solution (5 mg/mL) was added to each well for another culture for 4 h at 37°C. Then, 100 μL of DMSO was added into each well for 10 min. The optical density (OD) was measured by the microplate reader at 570 nm. Each assay was performed triplicate.
RNA Extraction and Microarray Analysis
The Agilent SurePrint G3 Human Gene expression v3 Microarrays (8*60K, Design ID: 072363) were used on the Agilent Microarray-Based Gene Expression Analysis platform by OEbiotech Co., Ltd. (Shanghai, P.R. China). In brief, the total RNA was extracted from MCF-7 cells treated with/without UA 20uM, and then quantified by the NanoDrop ND-2000 (Thermo Scientific) and the RNA integrity was assessed by Agilent Bioanalyzer 2100 (Agilent Technologies). The total RNA were reverse-transcribed to double-strand cDNA, then synthesized into cRNA and labeled with Cyanine-3-CTP. The labeled cRNAs were hybridized onto the microarray. After washing, the arrays were scanned by the Agilent Scanner G2505C (Agilent Technologies). The Feature Extraction software (Version10.7.1.1, Agilent Technologies) was used to analyze array images to get raw data. Genespring (Version 13.1, Agilent Technologies) was employed to finish the basic analysis with the raw data. To begin with, the raw data were normalized with the quantile algorithm. The probes that at least 1 condition out of 2 conditions has flags in “P” were chosen for further data analysis. Differentially expressed genes were then identified through fold change. The threshold set for up- and down-regulated genes was a fold change ≥ 2.0. Afterwards, GO analysis and KEGG analysis were applied to determine the roles of these differentially expressed mRNAs.
Western Blot
MCF-7 cells, incubated with (4 and 20 μM) UA for 24 h, were washed with cold phosphate buffer, then incubated with protein extract buffer for 30 min at −80°C. The lysates were centrifuged at 15,000 × g for 40 min and protein concentration was measured by BCA kit (Tiangen, Beijing, China). Protein lysates (25 μg) from each sample were subjected to SDS-PAGE on 15% acrylamide gel and transferred to the PVDF membrane. Blots were incubated with 5% nonfat milk to block nonspecific binding sites at 4°C overnight and then incubated with antibodies against BRAF, ERK1/2, IKKβ, NF-κB p65, PLK1, phosphor-ERK1/2, phosphor-p65, phosphor-IKKβ, phosphor-PLK1 and GAPDH for 1.5 h at 37°C. Membranes were then incubated with HRP-conjugated secondary antibodies for 1 h at 37°C. Finally, membranes were detected by ECL kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Molecular Docking
Docking prediction was performed with iGEMDOCK software. The structures of IKKβ (4KIK), BRAF (5CSW), and PLK1 (3RQ7) were obtained from the Protein Data Bank. The 3D structure of UA was obtained from National Center for Biotechnology Information, then transformed to MOL2 format using the software of Open Babel (v. 2.4.1). The binding sites were prepared, and energy-minimized compounds were imported. The docking protocol is set to 80 generations per ligand and a population size id 300 random individuals.
Statistics Analysis
Statistics analysis was performed with Origin 8.0. The data were expressed as mean±SD. Statistical analysis of the results was performed using the t-test for pairwise comparison. Differences were considered statistically significant at P<0.05 (*) or highly significant at p<0.01 (**).
Results
UA Inhibits Cell Viability of MCF-7 Cells
The inhibitory effect of UA on MCF-7 cell growth was determined by MTT assay. As shown in Figure 1B, the cell viability was decreased by UA in a dose-dependent manner. The IC50 values for UA to reduce cell viability were 20 μM (Figure 1B).
In addition, we found that 20 μM UA inhibited the growth of MCF-7 cells as early as 6 h (Supplementary Figure 1). Hence, the microarray and Western blot were performed on MCF-7 cells, treated with UA for 6 h and 24 h, respectively.
Microarray and CMAP Analysis of MCF-7 Treated with UA
MCF-7 cells, treated with 20 μM UA for 6 h, produced the initial gene expression profiles and fold change was used to filter the differential expression genes of UA (Supplementary Data 1). The result showed that it contained 355 up-regulated genes and 121 down-regulated genes (both >2-fold change, respectively). Then, the GO and KEGG pathway analyses were performed by using a multi-omics data analysis tool (Figure 2A and B).
The different gene expressions triggered by UA were submitted to the connectively MAP database for analysis. As shown in Table 1, the results suggested that UA exerted its anti-cancer activity through modulating RAF, PLK, IKK, and NF-κB, based on the character of BC. These findings were beneficial for exploring the anti-cancer mechanism of UA in MCF-7 cells.
 |
Table 1 The CMAP Analysis of the Different Gene Expression Triggered by UA |
UA Down-Regulates RAF/ERK Pathway
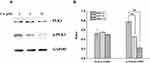
The CMAP results suggested that UA may act in a similar to that of RAF inhibitors. In BC, BRAF, the most important member of RAF, can activate ERK through phosphorylation to promote proliferation and invasion.19 Therefore, we evaluated the protein levels involved RAF/ERK pathway. As shown in Figure 3, UA treatment significantly reduced the phosphorylation levels of BRAF and ERK1/2 when compared to the untreated cells. The results suggest that UA exerts anti-BC effect via RAF/ERK pathway in MCF-7 cells.
UA Decreased the Phosphorylation Levels of PLK1
PLK (polo-like kinase), a family of serine/threonine phosphatase, is the key mediator of cancer cell growth and proliferation.20 Among the PLK members, PLK1 correlates with poor prognosis as the high expression in various cancers including BC.21,22 Hence, we investigated the effect of UA on PLK1 expression in MCF-7 cells. As shown in Figure 4, the phosphorylation level of PLK1 was significantly decreased by UA. These results were coordinated with the CMAP and microarray analysis and supported the notion that UA exerts an inhibition on the PLK phosphorylation.
UA Modulates IKK/NF-κB Pathway
The CMAP analysis indicated that UA has the similar effect on gene expression with NF-κB pathway inhibitor. In NF-κB pathway, IKKβ mediates phosphorylation of IκB/p65p50 complex. The IκB/p65p50 complex represents an inactivated cytoplasmic form of NF-κB.23 When IKKβ was activated, it phosphorylates the IκB/p65p50 complex, then releasing phosphorylated p65, which can interact with its DNA binding site to up-regulate the anti-apoptotic gene transcription, in cancer cells.24 To determine the effects of UA on the IKK/NF-κB pathway, the activation statuses of IKKβ and p65 were examined in MCF-7 cells. In Figure 5, UA treatment significantly reduced the phosphorylated levels of IKKβ and p65, without affecting total IKKβ and p65 expression. These results suggest that UA exerts an inhibition on the IKK/NF-kB pathway.
The Docking Analysis of UA with BRAF, IKK, and PLK1
iGEMDOCK is an important software for molecular docking, which predicts the mode of interaction between ligand and receptor. Binding energy provides the information to us for understanding the strength and how much affinity a compound binds to the pocket of the receptor. The docking results for iGEMDOCK showed that the binding energy of UA between BRAF, IKKβ, and PLK1 were −87.6, −99.7, and −78.2kcaj/mol, respectively. As shown in Figure 6, VAL-482, LYS-483, THR-529, TRP-531, CYS-532, and PHE-583, located in functional domain, were the amino acid residues in the binding sites of UA to BRAF (Figure 6A), while LEU-21, THR-23, VAL-29, CYS-99, ASP-103, and ILE-165 were responsible for UA binding to IKKβ (Figure 6B). For analyzing the binding sites between UA and PLK1, VAL-415, TYR-417, TYR-481, PHE-482, and TYR-485 were involved in stabilizing binding poses (Figure 6C).
Discussion
Previous studies have demonstrated that ursolic acid has the anti-tumor effect via mediating proliferation, apoptosis, and invasion in types of cancers, such as colorectal cancer,25 liver cancer,26 and squamous skin cancer,27 yet its underlying molecular mechanisms remain unresolved. In the present study, we explored the anti-tumor mechanism and potential targets of using CAMP database.
CMAP has been widely applied to drug discovery and pharmacological research. Tsang-Pai Liu et al identified two novel inhibitors (KM-00927 and BRD-K75081836) of histone deacetylase (HDAC).17 Our CMAP analysis showed that UA and IKK, PLK, and BRAF inhibitors had the obviously positive correlation in gene expression profiles by CMAP. Further data revealed that UA could inhibit PLK1, RAF/ERK, and IKK/NF-κB pathway by Western blot.
RAF/ERK pathway is the most frequently deregulated in cancer and plays a key role in carcinogenesis.28 Paramee et al showed that Kaempferia parviflora inhibits proliferation, migration, and induced apoptosis through down-regulating ERK pathway in ovarian cancer cells.29 Celastrus orbiculatus extracts induced apoptosis of gastric adenocarcinoma cell line MGC-803 via mediating ERK1/2.30 Our study showed that UA markedly suppressed the phosphorylation level of BRAF and ERK1/2.
The IKK/NF-κB pathway is also inhibited by UA. NF-κB is widely considered as a key mediator between inflammation and cancer. It regulates the transcriptional activation of genes with anti-apoptosis function, cell proliferation, tumor initiation, and progression, such as cyclin D1, Bcl-2, and so on.31–34 In the present study, we demonstrated that UA down-regulated the phosphorylation activation of IKKβ and p65, and docking analysis suggests that UA can bind to IKKβ function domain.
PLK1, which phosphorylate a myriad of centrosomal/mitotic protein substrates to insure the fidelity of mitotic progression.35 Donizy et al demonstrated that PLK1 displays an important role in breast progression.36 Zhang and Pati found that Sepin-1, a potent non-competitive inhibitor of separase, inhibits breast cancer cell growth through decreasing phosphorylation levels of RAF, PLK1, and FoxM1.37 The combination of PLK1 inhibitor and tankyrase-1 inhibitor could enhance the anti-cancer effect in triple-negative breast cancer cells.38 This study revealed the anti-BC mechanism of UA through the inhibition of PLK1 phosphorylation level and suggest that UA could interact with PLK1 structure in TYR-485, located in functional domain.
In conclusion, our results demonstrated that UA inhibited BC via modulating PLK1, IKK/NF-κB, and BRAF/ERK pathways. These findings provide a more comprehensive understanding of the mechanism by which UA exerts its anti-tumor effects and support its use in the treatment of breast cancer.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (grant no. 81502958), the National Natural Science Foundation of Jiangsu Province (grant no. BK20150286), and Basic research on the application of Suzhou (Agricultural Plans) (grant nos. SYN201506 and SYSD2017061).
Disclosure
The authors report no conflicts of interest in this work.
References
1. McGuire S. World cancer report 2014. Geneva, Switzerland: world Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr. 2016;7(2):418–419. doi:10.3945/an.116.012211
2. Siegel RL, Miller KD, Jemal A. Cancer statistic. CA Cancer J Clin. 2018;68:7–30. doi:10.3322/caac.21442
3. Fedele M, Cerchia I, Chiappetta G. The epithelial to mesenchymal transition in breast cancer: focus on basal like carcinomas. Cancers. 2017;9(10):134. doi:10.3390/cancers9100134
4. Iqbal J, Abbasi BA, Batool R, et al. Potential phytocompounds for developing breast cancer therapeutics: nature’s healing touch. Eur J Pharmacol. 2018;827:125–148. doi:10.1016/j.ejphar.2018.03.007
5. Gordaliza M. Natural products as leads to anticancer drugs. Clin Transl Oncol. 2007;9:767–776. doi:10.1007/s12094-007-0138-9
6. Nobili S. Natural compounds for cancer treatment and prevention. Pharmacol Res. 2009;59:365–378. doi:10.1016/j.phrs.2009.01.017
7. Cargnin ST, Gnoatto SB. Ursolic acid from apple pomace and traditional plants: a valuable triterpenoid with functional properties. Food Chem. 2017;220:477–489. doi:10.1016/j.foodchem.2016.10.029
8. Mancha-Ramirez AM, Slaga TJ. Ursolic acid and chronic disease: an overview of UA’s effects on prevention and treatment of obesity and cancer. Adv Exp Med Biol. 2016;928:75–96.
9. Chen CJ, Shih YL, Yeh MY, et al. Ursolic acid induces apoptotic cell death through AIF and endo G release through a mitochondria-dependent pathway in NCI-H292 human lung cancer cells in vitro. In Vivo. 2019;33(2):383–391. doi:10.21873/invivo.11485
10. Yang X, Li Y, Jiang W, et al. Synthesis and biological evaluation of novel ursolic acid derivatives as potential anticancer prodrugs. Chem Biol Drug Des. 2015;86:1397–1404.
11. Yeh CT, Wu CH, Yen GC. Ursolic acid, a naturally occurring triterpenoid, suppresses migration and invasion of human breast cancer cells by modulating c-Jun N-terminal kinase, AKT and mammalian target of rapamycin signaling. Mol Nutr Food Res. 2010;54(9):1285–1295. doi:10.1002/mnfr.200900414
12. Wang J, Liu L, Qiu H, et al. Ursolic acid simultaneously targets multiple signaling pathways to suppress proliferation and induce apoptosis in colon cancer cells. PLoS One. 2013;8(5):e63872. doi:10.1371/journal.pone.0063872
13. Lin Z, Jiang J, Liu XS. Ursolic acid mediated apoptosis of K562 cells involves Stat5/Akt pathway inhibition through the induction of Gfi-1. Sci Rep. 2016;6:33358. doi:10.1038/srep33358
14. Shishodia S, Majumdar S, Banerjee S, et al. Ursolic acid inhibits nuclear factor-κB activation induced by carcinogenic agents through suppression of IκBα kinase and p65 phosphorylation: correlation with down-regulation of cyclooxygenase 2, matrix metalloproteinase 9, and cyclin D1. Cancer Res. 2003;63(15):4375–4383.
15. Qu XYA, Rajpal DK. Applications of connectively map in drug discovery and development. Drug Discov Today. 2012;17:1289–1298. doi:10.1016/j.drudis.2012.07.017
16. Lamb J. The connectivity map: using gene-expression signatures to connect small molecules, genes and disease. Science. 2016;313:1929–1935.
17. Liu TP, Hsieh YY, Chou CJ, et al. Systematic polypharmacology and drug repurposing via an integrated L1000-based connectivity map database mining. R Soc Open Sci. 2018;5(11):181321. doi:10.1098/rsos.181321
18. Lee KH, Lo HL, Tang WC, et al. A gene expression signature-based approach reveals the mechanisms of action of the Chinese herbal medicine berberine. Sci Rep. 2014;4:6394. doi:10.1038/srep06394
19. Magdalena ZJ. The importance of autophagy regulation in breast cancer development and treatment. Biomed Res Int. 2014;2014:1–9.
20. Archambault V, Lepine G, Kachaner D. Understanding the polo kinase machine. Oncogene. 2015;34(37):4799–4807. doi:10.1038/onc.2014.451
21. Ueda A, Oikawa K, Fujita K, et al. Therapeutic potential of PLK1 inhibition in triple-negative breast cancer. Lab Invest. 2019;99(9):1275–1286. doi:10.1038/s41374-019-0247-4
22. Craig SN, Wyatt MD, McInnes C. Current assessment of polo-like kinases as anti-tumor drug targets. Expert Opin Drug Discov. 2014;9(7):773–789. doi:10.1517/17460441.2014.918100
23. Karin M, Cao Y, Greten FR, et al. NF-kappa B in cancer: form innocent bystander to major culprit. Nat Rev Cancer. 2002;2(4):301–310. doi:10.1038/nrc780
24. Park YH. The nuclear factor-kappa B pathway and response to treatment in breast cancer. Pharmacogenomics. 2017;18(18):1697–1709. doi:10.2217/pgs-2017-0044
25. Xavier CP, Lima CF, Pedro DF, et al. Ursolic acid induces cell death and modulates autophagy through JNK pathway in apoptosis-resistant colorectal cancer cells. J Nutr Biochem. 2013;24:706–712. doi:10.1016/j.jnutbio.2012.04.004
26. Liu T, Ma H, Shi W, et al. Inhibition of STAT3 signaling pathway by ursolic acid suppresses growth of hepatocellular carcinoma. Int J Oncol. 2017;51(2):555–562. doi:10.3892/ijo.2017.4035
27. Junco JJ, Mancha A, Malik G, et al. Resveratrol and P-glycoprotein inhibitors enhance the anti-skin cancer effects of Ursolic acid. Mol Cancer Res. 2013;11(12):1521–1529. doi:10.1158/1541-7786.MCR-13-0237
28. Yu Z, Ye S, Hu G, et al. The RAF-MEK-ERK pathway: targeting ERK to overcome obstacles to effective cancer therapy. Future Med Chem. 2015;7:269–289. doi:10.4155/fmc.14.143
29. Paramee S, Sookkhee S, Sakonwasun C, et al. Anti-cancer effects of Kaempferia parviflora on ovarian cancer SKOV3 cells. BMC Complement Altern Med. 2018;18(1):178. doi:10.1186/s12906-018-2241-6
30. Qian Y, Lu S, Shi Y, et al. Celastrus orbiculatus extracts induce apoptosis and inhibit invasion by targeting the maspin gene in human gastric adenocarcinoma cells. Oncol Lett. 2018;15:243–249. doi:10.3892/ol.2017.7341
31. Gilmore TD, Herscovitch M. Inhibitors of NF-κB signaling: 785 and counting. Oncogene. 2006;25:6887–6899. doi:10.1038/sj.onc.1209982
32. Xue W, Meylan E, Oliver TG, et al. Response and resistance to NF-κB inhibitors in mouse models of lung adenocarcinoma. Cancer Discov. 2011;1:236–247. doi:10.1158/2159-8290.CD-11-0073
33. González Quezada BA, Santana Bejarano U, Corona Rivera A, et al. Expression profile of NF-κB regulated genes in sporadic colorectal cancer patients. Oncol Lett. 2018;15:1–11.
34. Ren L, Li Z, Dai C, et al. Chrysophanol inhibits proliferation and induces apoptosis through NF-κB/cyclin D1 and NF-κB/Bcl-2 signaling cascade in breast cancer cell lines. Mol Med Rep. 2018;17:4376–4382. doi:10.3892/mmr.2018.8443
35. Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6(4):321–330. doi:10.1038/nrc1841
36. Donizy P, Halon A, Surowiak P, et al. Augmented expression of Polo-like kinase 1 is a strong predictor of shorter cancer-specific overall survival in early stage breast cancer at 15-year follow-up. Oncol Lett. 2016;12(3):1667–1674. doi:10.3892/ol.2016.4890
37. Zhang N, Pati D. Separase inhibitor sepin-1 inhibits foxm1 expression and breast cancer cell growth. J Cancer Sci Ther. 2018;10(3):517. doi:10.4172/1948-5956.1000517
38. Ha GH, Kim DY, Breuer EK, et al. Combination treatment of polo-like kinase 1 and tankyrase-1 inhibitors enhances anticancer effect in triple negative breast cancer cells. Anticancer Res. 2018;38(3):1303–1310. doi:10.21873/anticanres.12352
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.