Back to Archived Journals » International Journal of Interferon, Cytokine and Mediator Research » Volume 7
The -1082 A allele of the interleukin-10 single nucleotide polymorphism -1082 G/A is associated with hepatitis virus co-infection in Indonesian Javanese HIV patients
Authors Prasetyo AA, Nurfitria FB, Adnan ZA, Redhono D, Sari Y, Hudiyono
Received 12 February 2015
Accepted for publication 16 April 2015
Published 16 June 2015 Volume 2015:7 Pages 1—7
DOI https://doi.org/10.2147/IJICMR.S82759
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Randall Davis
Afiono Agung Prasetyo,1–3 Fadhila Balqis Nurfitria,1 Zainal Arifin Adnan,1,4 Dhani Redhono,4 Yulia Sari,1,2,5 Hudiyono1–3
1A-IGIC Research Group Faculty of Medicine, 2Center of Biotechnology and Biodiversity Research and Development, 3Department of Microbiology, 4Department of Internal Medicine, 5Department of Parasitology, Faculty of Medicine, Sebelas Maret University, Surakarta, Indonesia
Objective: This study was conducted to determine the distribution of the single nucleotide polymorphism (SNP) at position -1082 of the proximal promoter region of the interleukin (IL)-10 gene and its association with hepatitis virus co-infection in Indonesian Javanese human immunodeficiency virus (HIV)-infected patients.
Methods: Blood samples were collected from 156 Indonesian Javanese HIV-infected patients; tested with serological and molecular assays for hepatitis B virus (HBV), hepatitis C virus (HCV), and hepatitis D virus (HDV); and subjected to SNP -1082 G/A polymorphism analysis.
Results: Hepatitis B surface antigen (HBsAg) (+)/HBV DNA (+), HBsAg (-)/HBV DNA (+), anti-HCV antibodies (+)/HCV RNA (+), and anti-HCV antibodies (-)/HCV RNA (+) were detected from all of HIV-infected individuals at rates of 3.8% (6/156), 0.6% (1/156), 16.0% (25/156), and 11.5% (18/156), respectively. The frequencies of -1082 GG, -1082 GA, and -1082 AA were 14.7% (23/156), 60.3% (94/156), and 25% (39/156), respectively. HBV co-infection was found only in -1082 A allele carriers, and these patients also showed higher rates of HCV co-infection (odds ratio: 2.24, 95% confidence interval: 1.22–4.10, P=0.009).
Conclusion: The -1082 A allele was associated with HBV and HCV co-infection in Indonesian Javanese HIV patients.
Keywords: IL-10 -1082, HIV, HBV, HCV, Indonesia
Introduction
Human immunodeficiency virus (HIV) co-infection with hepatitis B virus (HBV) or hepatitis C virus (HCV) has become a major concern worldwide. Approximately 10% of people infected with HIV are estimated to be co-infected with HBV, and 15%–30% are estimated to be co-infected with HCV.1,2 In Indonesia, HIV/HBV co-infection rates reportedly range from 0% to 27.1%, while HIV/HCV rates range from 4% to 34.1%.3–6 HIV, HBV, and HCV infections are persistent, and co-infection by these pathogens amplifies the burden of disease.1,2,7
The persistence of HIV, HBV, and HCV infections has been associated with immunosuppression during the initial immune response.7 Interleukin (IL)-10, a cytokine capable of indirect repression of the T helper 1 (Th1) response, is a key player in the immunosuppression that directly leads to viral persistence.8,9 IL-10 can be induced by pathogen or host factors, and the levels of IL-10 produced may vary among individuals.10,11
A single nucleotide polymorphism (SNP) at position -1082 of the proximal promoter region of the IL-10 gene (rs1800896) has been associated with differing levels of IL-10 production across three genotypes (-1082 GG, -1082 GA, and -1082 AA).12 High production of IL-10 is associated with the -1082 GG genotype; low production of IL-10 is associated with the -1082 AA genotype; and IL-10 production in the -1082 GA genotype falls between the two extremes.12 The -1082 AA genotype has also been associated with HIV type 1 (HIV-1) acquisition and cluster differentiation 4+ (CD4+) Th cell decline acceleration rates in late stages of the disease.13,14 In the acute phase of HIV-1 infection, IL-10 levels may give way to viral replication by dampening the innate and adaptive immune responses, whereas in chronic HIV-1 infection, IL-10 may be protective by reducing immune activation and inhibiting HIV-1 replication in macrophages.15 The IL-10 SNP -1082 has also repeatedly been associated with HBV and HCV infections.16–21 However, no study has been performed to assess the association of IL-10 SNP -1082 with hepatitis virus co-infection in HIV-infected patients. Moreover, data on IL-10 SNP -1082 allele frequencies in Indonesians are lacking.
Materials and methods
Study population
Since 2009, our research group has conducted a molecular epidemiology study of human blood-borne viruses by collecting epidemiological data and blood samples from high-risk communities in the Central Java province, Indonesia. From November 2011 to February 2012, Indonesian Javanese adult HIV patients in the Voluntary Counseling and Testing Clinic at the Dr Moewardi General Hospital in Surakarta (n=156) were enrolled in the study. Blood samples collected from these patients were subjected to hematological and CD4+ T cells assays, fractionated, aliquoted, and kept frozen until analysis. Approval was obtained from the institutional ethical committee review boards of the Faculty of Medicine of Sebelas Maret University and Dr Moewardi General Hospital, Surakarta, Indonesia. Written informed consent was obtained from all individuals participating in the study. All procedures were conducted according to the principles of the Declaration of Helsinki.
Blood sample collection and screening for infection
Blood samples were obtained from each participant in ethylene diamine-tetra-acetic acid (EDTA) tubes. The blood samples were also screened for HBV, HCV, and hepatitis D virus (HDV) by serological assays, as described previously.5 Briefly, a SERATEC Hepatitis B Quick Test (Gesellschaft für Biotechnologie GmbH, Göttingen, Germany) was used to detect hepatitis B surface antigen (HBsAg). Ortho HCV PA II (Ortho Diagnostics, Tokyo, Japan) and HDV Ab ELISA kits (Diagnostic Automation, Calabasas, CA, USA) were used to detect anti-HCV and anti-HDV antibodies, respectively. All assays were performed according to the manufacturer’s instructions. All samples were tested in duplicate at minimum.
Nucleic acid extraction and molecular detection
Viral nucleic acid extraction and molecular detection were performed as described previously.5 Briefly, nucleic acid (DNA and RNA) was extracted from 200 μL of each plasma sample using the PureLink Viral RNA/DNA Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The nucleic acids were then aliquoted, and one aliquot was reverse-transcribed according to the Superscript III First-Strand cDNA Synthesis Supermix Kit protocol using random hexamers (Invitrogen). Molecular detection was performed by polymerase chain reaction (PCR) using the AmpliTaq Gold® 360 DNA Polymerase Kit (Invitrogen). A portion of the HIV gag gene encoding the p24 region was amplified. A portion of the HBsAg gene was amplified by nested PCR. A portion of the NS5B region and a portion of the E1–E2 region, including hyper variable region-1 of the HCV genome, were amplified by nested PCR. HDV RNA was detected by the amplification of a 400-nucleotide (nt)-long region of the HDV genome proposed for the classification of HDV genotypes. Internal amplification controls were included to exclude any false-negative results. The corresponding positive controls and one negative control (sterile water) were included for each group simultaneously. To prevent PCR contamination, the reagent preparation, sample processing, and nested PCR assays were performed in rooms separate from those where the amplified products were analyzed. Aerosol-resistant pipette tips were used throughout the assays. The PCR products were subjected to electrophoresis in 2% agarose gels, which were stained with ethidium bromide and visualized under ultraviolet illumination. The specificity was confirmed by sequencing the amplicons. All samples were tested in duplicate at minimum.
Detection of SNP -1082 G/A
Genomic DNA from each patient was isolated using the High Pure PCR Template Preparation Kit (Roche Diagnostics, Mannheim, Germany). For SNP -1082 G/A polymorphism analysis, genomic samples were genotyped by PCR using the FastStart HiFi PCR System dNTPack (Roche Diagnostics), with primer pairs previously described.22 The PCR products were subjected to electrophoresis in 2% agarose, stained with ethidium bromide, and visualized under ultraviolet illumination. To avoid false negatives, an internal control primer, to amplify a 429-bp region of the human growth hormone (HGH) gene, was used for all PCR reactions.22 All samples were tested twice at minimum.
IL-10 plasma measurements
The IL-10 levels in the plasma samples were measured using a Human IL-10 Immunoassay Quantikine ELISA (R&D Systems, Minneapolis, MN, USA). The cytokine concentration in the samples was calculated in pg/mL using recombinant human IL-10 (R&D Systems) as a standard. The absorbance was measured at 450 nm with an ELISA plate reader, and the reading at 540 nm was subtracted to correct for optical imperfections in the plate. All samples were tested in triplicate at minimum to calculate the mean level of IL-10 in each sample.
Statistical analysis
Statistical analysis was performed using SPSS version 20 software (SPSS, Chicago, IL, USA). A 95% confidence interval (CI) was used for all data analysis. Independent sample t-tests and Mann–Whitney U tests were used to compare the means of the data. Chi-square or Fisher’s exact test, as indicated, was used to compare qualitative data. P-values less than 0.05 were considered significant.
Results
General characteristics of the study population
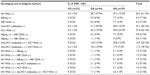
This study included 156 Indonesian Javanese HIV-infected patients, whose general characteristics are tabulated in Table 1. The samples were derived from 86 males (55.1%) and 70 females (44.9%). The mean age of the patients was 36.9±8.7 years, ranging from 19 years to 72 years. Most of the patients received combination of zidovudine, lamivudine, and nevirapine as standard regimen of antiretroviral therapy (ART) in the Voluntary Counseling and Testing Clinic at the Dr Moewardi General Hospital in Surakarta. Twenty of patients (8 males, 12 females) reported that they had not started ART. The levels of IL-10 were not significantly different between the antiretroviral-treated and untreated patients (P=0.782). Eosinophilia was found predominantly in male patients (odds ratio [OR]: 2.49, 95% CI: 1.30–4.76, P=0.006). Unless otherwise stated, no statistical associations could be drawn.
HIV and hepatitis co-infection in patients
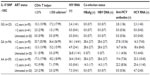
HIV RNA was detected in 26.3% (41/156) of the samples (Table 2), more frequent (70.0%, 14/20) in antiretroviral-untreated patients (OR: 9.42, 95% CI: 3.31–26.79, P<0.001), or in antiretroviral-treated for 2 years or less (26.3%, 25/95, OR: 6.96, 95% CI: 1.57–30.98, P=0.011) (Table 3). HBsAg (+)/HBV DNA (+) and HBsAg (–)/HBV DNA (+) results were detected in 3.8% (6/156) and 0.6% (1/156) of the samples, respectively (Table 2). The anti-HCV antibody positive rate was 23.1% (36/156), and this finding was significantly more predominant in male patients (OR: 2.20, 95% CI: 0.99–4.87, P=0.049). The HCV RNA positive rate was 27.6% (43/156) and also occurred more frequently in male patients (OR: 1.54, 95% CI: 0.75–3.17, P=0.235). Patients with elevated IL-10 were more likely to present with HCV co-infection, indicated by the presence of HCV RNA (OR: 2.76, 95% CI: 1.26–6.04, P=0.010). HDV was not detected in any patient.
Genotypic distribution and hematologic findings for IL-10 SNP -1082
In our study population, the frequency of the -1082 GA genotype was the highest (60.3%, 94/156), followed by that of the -1082 AA (25%, 39/156) and -1082 GG genotypes (14.7%, 23/156), respectively, which defied Hardy–Weinberg equilibrium (P=0.007). Leukocytopenia (OR: 1.38, 95% CI: 0.83–2.30, P=0.215), lymphocytopenia (OR: 1.10, 95% CI: 0.65–1.86, P=0.725), a CD4+ Th cell frequency >21% (OR: 1.05, 95% CI: 0.63–1.74, P=0.862), and a CD4+ Th cell count >350 cells/mm3 (OR: 1.03, 95% CI: 0.62–1.71, P=0.918) were frequently found in the -1082 A allele carriers. The mean percentage and absolute counts of CD4+ Th cells in patients with the -1082 GG genotype (14.6%, standard deviation 7.7; 262.0 cells/mm3, standard deviation 154.6) were lower (P<0.001) compared with that in the other genotypes. IL-10 plasma levels were not associated with the genotype variations of IL-10 SNP -1082.
Associations between IL-10 SNP -1082 genotypes and HIV/HBV and HIV/HCV co-infections
HIV RNA was detected more frequent in -1082 A allele carriers (-1082 GA and AA) (OR: 1.37, 95% CI: 0.78–2.40, P=0.277, Table 2). HBV co-infection was only found in -1082 A allele carriers. One patient with the -1082 GA genotype was HBsAg-negative but HBV DNA-positive. Anti-HCV antibodies (OR: 1.63, 95% CI: 0.89–3.00, P=0.115) and HCV RNA (OR: 2.24, 95% CI: 1.22–4.10, P=0.009) were also predominantly found in -1082 A allele carriers. Anti-HCV antibody (+)/HCV RNA (+) results were detected only in -1082 A allele carriers. To confirm the present finding, we tested the Indonesian Javanese “hepatitis virus positive only” samples collected from our previous study.4,5 In those samples, HBsAg positive (n=23) and HBV DNA (n=20) were only found in -1082 allele carriers. Anti-HCV antibodies (53.2%, 149/280) and HCV RNA (19.6%, 55/280) were also predominantly found in -1082 A allele carriers (OR: 1.61, 95% CI: 1.11–2.34, P=0.012 and OR: 2.14, 95% CI: 1.17–3.91, P=0.014, respectively) in those samples.
Discussion
In our study, the -1082 GA genotype was found in the highest frequency, followed by -1082 AA and GA, which defied Hardy–Weinberg equilibrium. This finding also differs from a previous study involving a Thai population, in which -1082 AA was found in the highest frequency (87.3%).23 Moreover, the -1082 AA genotype frequency was reported to be extremely high in Japanese (92.4%) and Chinese populations (92.3%).17,21 Ethnic differences between study populations may be one of the reasons for these distinctive results. However, our results contribute additional information regarding the distribution of SNP -1082 alleles in Asia, particularly Southeast Asia, as data on the distribution of SNP -1082 genotypes across ethnic groups in this region remain extremely limited.
The IL-10 SNP -1082 polymorphism was analyzed for its association with the most common biomarker of disease progression, CD4+ Th cell levels. The mean CD4+ Th cell percentages in patients with the -1082 GG genotype were significantly lower than those in -1082 A allele carriers (-1082 GA and -1082 AA), consistent with a prior study.24 Thus, the IL-10 SNP -1082 may have underlying but as-yet-undetermined epigenetic or environment modulatory factors, or this SNP may be in linkage disequilibrium (LD) with other genes, with a modulatory effect on attenuating CD4+ T cell decline.24
The levels of plasma IL-10 were not associated with the -1082 SNP genotypes (GG, GA, and AA), consistent with a previous study.25 However, the elevation of IL-10 was associated with HIV/HCV co-infection, indicated by the presence of HCV RNA, and is also consistent with a previous study.26 Upregulation of IL-10 expression has been associated with the interaction between the HCV core protein and Toll-like receptor-2 and has been shown to be induced by non-structural protein 3 in macrophages and dendritic cells.26,27 Moreover, IL-10 was significantly associated with preserved levels of CD4 T cells in HIV patients, indicating that HCV co-infection may be contributing to unbalanced persistent inflammation.28
Previously, the -1082 GG genotype was reported as a host genetic factor in the susceptibility to HBV infection in the Egyptian population.29 However, in a meta-analysis performed by Lu et al, the G allele of the IL-10 SNP -1082 was not associated with HBV infection in the Asian population.30 In the present study, all HBV co-infections were found only in -1082 A allele carriers (-1082 AA and -1082 GA), suggesting that the -1082 A allele is associated with HBV infection, as reported previously in non-HIV-infected patients.17,31,32 This same polymorphism may play a different role in disease susceptibility in different ethnic populations, as different populations often have different LD patterns.29 However, HCV co-infections were also found to be predominant in -1082 A allele carriers, consistent with previous studies performed in HCV-mono-infected Chinese populations,33,34 suggesting a potential role for the -1082 allele in hepatitis virus susceptibility in Indonesian Javanese patients.
Conclusion
In summary, -1082 A allele carriers (-1082 GA and AA), particularly Indonesian Javanese patients, showed increased rates of HIV/HBV and HIV/HCV co-infections. Although this study focused on a relatively small number of individuals, these findings contribute to evidence of the presence of genetic variants important for predicting host susceptibility to hepatitis virus co-infection in HIV patients.
Acknowledgments
This work was supported in part by grants from the Indonesian Directorate of Higher Education (No 505/SP2H/PL/Dit.Litabmas/VII/2011, No 197/SP2H/PL/Dit.Litabmas/IV/2012, and No 077/SP2H/PL/Dit.Litabmas/V/2013) and from APBN/DIPA UNS (No 2342/UN27.16/PN/2012, No 159a/UN27.11/PN/2013, No 165/UN27.11/PN/2013, No 267a/UN27.16/PN/2013, No 501/UN27.11/PN/2014, No 853/UN27.11/PN/2014, and No 339/UN27.11/PL/2015).
Disclosure
The authors report no conflicts of interest in this work.
References
Ranjbar R, Davari A, Izadi M, Jonaidi N, Alavian SM. HIV/HBV co-infections: epidemiology, natural history, and treatment: a review article. Iran Red Crescent Med J. 2011;13(12):855–862. | |
Andreoni M, Giacometti A, Maida I, Meraviglia P, Ripamonti D, Sarmati L. HIV-HCV co-infection: epidemiology, pathogenesis and therapeutic implications. Eur Rev Med Pharmacol Sci. 2012;16(11):1473–1483. | |
Anggorowati N, Yano Y, Heriyanto DS, et al. Clinical and virological characteristic of hepatitis B or C virus co-infection with HIV in Indonesia patients. J Med Virol. 2013;84(6):857–865. | |
Prasetyo AA, Ariapramuda ER, Al Kindi E, et al. Men having sex with men in Surakarta, Indonesia: demographics, behavioral characteristics and prevalence of blood borne pathogens. Southeast Asian J Trop Med Public Health. 2014;45(5):1032–1047. | |
Prasetyo AA, Dirgahayu P, Sari Y, Hudiyono H, Kageyama S. Molecular epidemiology of HIV, HBV, HCV, and HTLV-1/2 in drug abuser inmates in central Java prisons, Indonesia. J Infect Dev Ctries. 2013;7(6):453–467. | |
Utsumi T, Yano Y, Lusida MI, et al. Detection of highly prevalent hepatitis B virus co-infection with HIV in Indonesia. Hepatol Res. 2013;43(10):1032–1039. | |
Wilson E, Brooks D. The role of IL-10 in regulating immunity to persistent viral infections. Curr Top Microbiol Immunol. 2011;350:39–65. | |
Brockman MA, Kwon DS, Tighe DP, et al. IL-10 is upregulated in multiple cell types during viremic HIV infection and reversibly inhibits virus-specific T cells. Blood. 2009;114(2):346–356. | |
Brooks DG, Walsh KB, Elsaesser H, Oldstone MB. IL-10 directly suppresses CD4 but not CD8 T cell effector and memory responses following acute viral infection. Proc Natl Acad Sci U S A. 2010;107:3018–3023. | |
Reuss E, Fimmers R, Kruger A, Becker C, Rittner C, Höhler T. Differential regulation of interleukin-10 production by genetic and environmental factors – a twin study. Genes Immun. 2002;3(7):407–413. | |
Mörmann M, Rieth H, Hua TD, et al. Mosaics of gene variations in the Interleukin-10 gene promoter affect interleukin-10 production depending on the stimulation used. Genes Immun. 2004;5(4):246–255. | |
Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hitchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24(1):1–8. | |
Erikstrup C, Kallestrup P, Zinyama-Gutsire RB, et al. Reduced mortality and CD4 cell loss among carriers of the interleukin-10 -1082G allele in a Zimbabwean cohort of HIV-1-infected adults. AIDS. 2007;21(17):2283–2291. | |
Shin HD, Winkler C, Stephens JC, et al. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL-10. Proc Natl Acad Sci U S A. 2000;97(26):14467–14472. | |
Naicker DD, Werner L. Interleukin-10 promoter polymorphisms influence HIV-1 susceptibility and primary HIV-1 pathogenesis. J Infect Dis. 2009;400(3):448–452. | |
Chuang JY, Yang SS, Lu YT, et al. IL-10 promoter polymorphisms and sustained response to combination therapy in Taiwanese chronic hepatitis C patients. Dig Liver Dis. 2009;41(6):424–430. | |
Migita K, Miyazoe S, Maeda Y, et al. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection – association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol. 2005;42(4):505–510. | |
Paladino N, Fainboim H, Theiler G, et al. Gender susceptibility to chronic hepatitis C virus infection associated with Interleukin 10 promoter polymorphism. J Virol. 2006;80(18):9144–9150. | |
Pár A, Pár G, Tornai I, et al. IL28B and IL-10R -1087 polymorphisms are protective for chronic genotype 1 HCV infection and predictors of response to interferon-based therapy in an East-Central European cohort. BMC Res Notes. 2014;7:12. | |
Truelove A, Oleksyk T. Evaluation of IL-10, IL19 and IL20 gene polymorphisms and chronic hepatitis B infection outcome. Int J Immunogenet. 2008;35(3):255–264. | |
Wang S, Huang D, Sun S, Ma W, Zhen Q. Interleukin-10 promoter polymorphism predicts initial response of chronic hepatitis B to interferon alfa. Virol J. 2011;8(1):28. | |
Korachi M, Pravica V, Barson AJ, Hutchinson IV, Drucker DB. Interleukin 10 genotype as a risk factor for sudden infant death syndrome: determination of IL-10 genotype from wax-embedded postmortem samples. FEMS Immunol Med Microbiol. 2004;41(1):125–129. | |
Kingkeow D, McNicholl JM, Maneekarn N, Wongtrakul J. Frequencies of IL-10 SNP genotypes by multiplex PCR-SSP and their association with viral load and CD4 counts in HIV-1-infected Thais. Asian Pac J Allergy Immunol. 2011;29(1):94–101. | |
Naicker DD, Wang B, Losina E, et al. Association of IL-10-promoter genetic variants with the rate of CD4 T-cell loss, IL-10 plasma levels, and breadth of cytotoxic T-cell lymphocyte response during chronic HIV-1 infection. Clin Infect Dis. 2012;54(2):294–302. | |
Talaat RM, Ashour ME, Bassyouni IH, Raouf AA. Polymorphisms of interleukin 6 and interleukin 10 in Egyptian people with Behcet’s disease. Immunobiology. 2014;219(8):573–582. | |
Dolganiuc A, Chang S, Kodys K, et al. Hepatitis C virus (HCV) Core protein-induced, monocyte-mediated mechanisms of reduced IFN-alpha and plasmacytoid dendritic cell loss in chronic HCV infection. J Immunol. 2006;177(10):6758–6768. | |
Dolganiuc A, Kodys K, Kopasz A, et al. Hepatitis C Virus Core and nonstructural protein 3 proteins induce pro- and anti-inflammatory cytokines and inhibit dendritic cell differentiation. J Immunol. 2003;170(110):5615–5624. | |
Villacres MC, Kono N, Mack WJ, et al. Interleukin 10 responses are associated with sustained CD4 T cell counts in treated HIV infection. J Infect Dis. 2012;206(5):780–789. | |
Talaat RM, Dondeti MF, El-Shenawy SZ, Khamiss OA. Association between IL-10 gene promoter polymorphism and hepatitis B viral infection in an Egyptian population. Biochem Genet. 2014;52(9–10):387–402. | |
Lu YL, Wu X, Huang HL, Dai LC. Allele polymorphisms of interleukin-10 and hepatitis B, C virus infection. Chin Med J (Engl). 2010;123(10):1338–1344. | |
Zhang TC, Pan FM, Zhang LZ, et al. A meta-analysis of the relation of polymorphism at sites -1082 and -592 of the IL-10 gene promoter with susceptibility and clearance to persistent hepatitis B virus infection in the Chinese population. Infection. 2011;39(1):21–27. | |
Shin HD. Interleukin 10 haplotype associated with increased risk of hepatocellular carcinoma. Hum Mol Genet. 2003;12(8):901–906. | |
Gao QJ, Liu DW, Zhang SY, et al. Polymorphisms of some cytokines and chronic hepatitis B and C virus infection. World J Gastroenterol. 2009;15(44):5610–5619. | |
Ishida C, Ikebuchi Y, Okamoto K, Murawaki Y. Functional gene polymorphisms of interleukin-10 are associated with liver disease progression in Japanese patients with hepatitis C virus infection. Intern Med. 2011;50(7):659–666. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.