Back to Journals » OncoTargets and Therapy » Volume 12
THAP7 promotes cell proliferation by regulating the G1/S phase transition via epigenetically silencing p21 in lung adenocarcinoma
Authors Chen CP, Sang Y, Liu L, Feng ZQ, Liang Z , Pei X
Received 15 March 2019
Accepted for publication 3 July 2019
Published 12 July 2019 Volume 2019:12 Pages 5651—5660
DOI https://doi.org/10.2147/OTT.S208908
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Cai-Ping Chen,1,* Yi Sang,2,* Lijuan Liu,3,* Zhi-Qi Feng,1 Zibin Liang,4 Xiaofeng Pei4
1Jiangsu Key Laboratory of Drug Discovery for Metabolic Disease, and State Key Laboratory of Natural Medicines, China Pharmaceutical University, Nanjing 210009, People’s Republic of China; 2Department of Center Laboratory, Jiangxi Key Laboratory of Cancer Metastasis and Precision Treatment, The Third Affiliated Hospital of Nanchang University, Nanchang, Jiangxi 330008, People’s Republic of China; 3Department of Pharmacy, Jiangxi Cancer Hospital, Nanchang, Jiangxi 330029, People’s Republic of China; 4Department of Thoracic Oncology, The Cancer Center of the Fifth Affiliated Hospital of Sun Yat-sen University, Zhuhai, Guangdong 519000, People’s Republic of China
*These authors contributed equally to this work
Purpose: Lung adenocarcinoma (LUAD) is one of the most common cancers worldwide. The THanatos-Associated Proteins (THAP) family plays an essential role in multiple cancers. However, the role of THAP7 in cancers has remained elusive.
Methods: THAP7 expression status in LUAD tissues was analysed by using the Oncomine database and qRT-PCR, and its expression level in LUAD cell lines was detected by qRT-PCR and Western blotting. The role of THAP7 in LUAD cells was determined by proliferation, colony formation, and cell cycle analyses. In vivo role of THAP7 was studied on xenograft models. Luciferase reporter assays and chromatin immunoprecipitation (ChIP) were used to determine the activity and acetylation of the p21 promoter.
Results: THAP7 expression was increased in LUAD tissues and cell lines. Moreover, the high expression of THAP7 was correlated with poor prognosis. The overexpression of THAP7 accelerated the G1/S phase transition and promoted tumour growth both in vitro and in vivo. A mechanistic study revealed that THAP7 reduced the acetylation of histone H3 on the p21 promoter to suppress p21 transcription.
Conclusion: For the first time, we demonstrated the function of THAP7 in LUAD, and our findings suggested that THAP7 may be a potential molecular therapy target in LUAD.
Keywords: THAP7, lung adenocarcinoma, proliferation, cell cycle, p21
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide and is associated with an overall 5-year survival rate lower than 16%.1–4 Non-small cell lung carcinoma (NSCLC) accounts for approximately 75–85% of the total number of lung cancers, of which adenocarcinoma is the main subtype.5,6 Most cases of lung adenocarcinoma, often without obvious clinical symptoms at the early diagnosis, are generally diagnosed with locally advanced or metastatic diseases.7 Moreover, lung adenocarcinoma was not sensitive to conventional chemotherapy. Therefore, a better understanding of the molecular mechanisms of lung adenocarcinoma pathogenesis and the identification of novel therapeutic targets are urgently needed.
The THanatos-Associated Proteins (THAPs) were discovered at the beginning of the 2000s.8,9 A total of twelve human THAP proteins (THAP0-11) have been identified so far. The THAP signature consists of a C2CH module that provides ligands for zinc coordination, four invariant hydrophobic residues and a C-terminus AVPTIF motif that is relatively well conserved within the THAP family.10 The THAP domain allows these proteins to bind to their DNA targets through the formation of multi-protein complexes and the transcription regulation of gene subsets.11 THAP-containing proteins serve as transcriptional regulators linked to cell proliferation, cell cycle progression, mitochondrial function, maintenance of pluripotency, angiogenesis, apoptosis and epigenetic gene silencing.12–15 Moreover, the deregulation of human THAP proteins has been associated with severe human diseases, including several types of cancers.16–18 Thanatos-associated protein-7 (THAP7), a member of the THAP family, was first shown to bind to hypoacetylated histone H4 tails via its C-terminal 77 amino acids.19 THAP7 represses transcription by recruiting NCoR/HDAC3 and TAF-Iβ and HiNF-P.19–21 However, to our knowledge, the role of THAP7 in cancers has not been reported.
In this study, we characterized THAP7 as a tumour promoter in LUAD cells. Moreover, we verified that THAP7 suppressed p21 expression via the deacetylation of histone H3 on its promoter.
Materials and methods
Patients, tissue specimens, and cell lines
Fourteen pairs of LUAD tissues and corresponding noncancerous adjacent tissues were obtained from patients diagnosed with NSCLC at the Fifth Affiliated Hospital of Sun Yat-sen University. All the involved patients were informed and consent was written and collected. The study was approved by the Clinical Research Ethics Committees of the Fifth Affiliated Hospital of Sun Yat-sen University (No 20190136-L).
Three human lung cancer cell lines (H1299, H1975, and A549) and the human bronchial epithelial cell line BEAS-2B were purchased from the American Type Culture Collection (ATCC) and cultured according to the instructions. The four cell lines used in this study were authenticated by short-tandem repeat analysis, and all cells were cultured within 2 months after thawing.
Vector construction
THAP7 overexpression plasmid was obtained by cloning full-length THAP7 cDNA into the pSin-puro vector. The lentiviral transfer plasmid carrying THAP7 shRNA was established using the Sigma-Aldrich shRNA system (Merck KGaA) according to the manufacturer’s instructions. A luciferase reporter vector was generated by cloning the promoter of p21 into the pGL3-basic vector. All recombinant plasmids were verified by DNA sequencing.
Establish stable cell lines
The lentiviral transfer plasmid delivering THAP7 or shTHAP7 was co-transfected with pMD.2G and psPAX2 into HEK-293T cells for 48 h to allow packaging. The recombinant viruses were subsequently collected and infected into A549 cells or H1299 cells in the presence of 8 µg/ml polybrene. The stable lines were obtained after puromycin (1 µg/ml) selection.
RNA extraction and quantitative real-time (qRT-PCR)
The procedures were performed as previously described.4 Briefly, total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. First-strand cDNA was synthesized using the Revert AidTM First Strand cDNA Synthesis Kit (MBI Fermentas). The primers employed for amplifying THAP7, p21 and GAPDH were validated. The primers used are as follows: THAP7, 5’-CAAGACCAAAGGACACAGTTACC-3’ (for) and 5’-GTCTGAAAAGGGGCT GCTAAG-3’ (rev). p21, 5’-CGATGGAACTTCGACTTTGTCA-3’ (for) and 5’-GCACAAGGGTACAAGACAGTG-3’ (rev). GAPDH, 5’-ACAGTCAGCCGCATCTTCTT-3’ (for) and 5’-GACAAGCTTCCCGTTCTCAG-3’ (rev).
Western blotting
Total protein from cultured cells was extracted with RIPA buffer containing protease and phosphatase inhibitors. Equal amounts of protein were subjected to Western blotting as described previously.4,22,23 The primary antibodies used were as follows: anti-THAP7 was purchased from Sigma-Aldrich, anti-p21 was obtained from Cell Signalling Technology (Cambridge, MA, USA), and anti-GAPDH was purchased from Bioworld Company (Nanjing, China).
Cell proliferation
Cells were seeded in 96-well plates at a density of 1,000 cells/well and grew for 1, 2, 3, 4, or 5 days. Then, 10 µL of the CCK-8 reagent (Cell Counting Kit-8, Beyotime, China) was added to each well, followed by incubation for 1.5 h. The absorbance at 450 nm was then recorded. For each experimental condition, 6 wells were used.
Colony formation assay
Cells were plated into 6-well plates at 500 cells/well and allowed to culture for 15 days at 37 °C. Then, the cells were stained with Giemsa solution after been washed twice with PBS. The number of colonies containing ≥50 cells was counted under a microscope.
Luciferase assay
The assay was carried out as described previously.24,25 Briefly, A549 cells were seeded onto 12-well plates at a density of 3×105/well and transfected with 0.8 µg promoter-luciferase plasmid. To normalize transfection efficiency, the cells were co-transfected with 8 ng pRL-CMV encoding Renilla luciferase. After transfection for 48 h, luciferase activity was measured using a dual-luciferase assay kit (Promega, Madison, WI, USA). Three independent experiments were performed.
Animal experiments
All animal studies were performed in accordance with protocols approved by Nanchang University (Nanchang, China). The mice were maintained in specific pathogen-free conditions at a temperature of 20–25 °C and 50–70% humidity under a light/dark cycle of 12 h, with free access to water and food. A total of 12 4-week-old male athymic nude mice were obtained from Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). For subcutaneous injection, 1×106 cells were mixed with 0.2 ml PBS (pH 7.4) and 30% (v/v) Matrigel matrix (BD Biosciences). The suspensions were injected subcutaneously into the flanks of 4-week-old male athymic nude mice. Tumours were excised and weighted after 18 days.
Chromatin immunoprecipitation (ChIP) assay
These procedures were performed as previously described.3,24,25 Briefly, the ChIP assay was performed using a ChIP kit (cat. no. 53008, Active Motif, Carlsbad, CA, USA) as described previously. Briefly, to fix the cells, Complete Cell Fixative Solution (included in kit) was added to the existing culture medium for the cells at 80% confluence at room temperature, and the fixation reaction was stopped by adding Stop Solution (included in kit) to the existing culture medium. The cells were collected by centrifugation at 1,000×g for 5 min at 4 °C. Subsequently, the nuclear pellet was resuspended in ChIP Buffer (included in the kit). The cell lysate was subjected to shearing using a sonication instrument (Ningbo Scientz Biotechnology Co., Ltd., Ningbo, China) to a fragment length of 200–500 bp. Total genomic DNA (input) was quantified, and 20 µg chromatin from each sample was immunoprecipitated overnight at 4 °C with 5 µg anti-H3ac (pan-acetyl) or normal IgG as a negative control. Then, nucleosome complexes were isolated with the protein G agarose beads for 3 h at 4 °C. Bound DNA-protein complexes were eluted, and cross-links were reversed after a series of washes using the washing reagent contained in the ChIP kit. Purified DNA was resuspended in TE buffer. Subsequently, the PCR was performed using PrimeSTAR® Max DNA Polymerase (cat. no. R045A, Takara Bio, Inc.). Thermal cycling of the qPCR reaction was initiated with a denaturation step at 94 °C for 2 min, followed by 35 cycles of denaturation at 98 °C 10 sec, annealing at 60 °C for 15 sec and elongation at 72 °C for 30 sec. The primers for ChIP are as follows: 5’-GGTGTCTAGGTGCTCCAGGT-3’ (for) and 5’-GACACAGCACTGTTAGAATG AGCC-3’ (rev).
Public data acquisition
The public Oncomine database (https://www.oncomine.org/resource/login.html) was used to explore THAP7 expression in lung adenocarcinoma. The correlations of THAP7 mRNA levels with the overall survival of lung adenocarcinoma patients were analysed using the online KMplot database (http://www.KMplot.com).
Statistical analysis
All statistical analyses were performed using SPSS for Windows, version 16.0 (SPSS). All in vitro data are expressed as the mean ± SD from at least three independent experiments. Student’s t test was used to estimate the difference between two groups. The Mann–Whitney test was used to assess the expression of THAP7 in lung adenocarcinoma tissues and normal tissues. Survival was analysed using the Kaplan–Meier method with a log-rank test. A p-value<0.05 was considered statistically significant.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Clinical Research Ethics Committees of the Fifth Affiliated Hospital of Sun Yat-sen University and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The animal studies were in accordance with the Animal Ethics Committee of Nanchang University.
Results
THAP7 expression is upregulated in lung adenocarcinoma
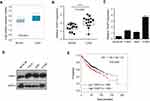
To explore the role of THAP7 in the progression of lung adenocarcinoma, we first analysed its expression status in lung adenocarcinoma using the Oncomine database. As shown in Figure 1A, THAP7 was significantly upregulated in lung adenocarcinoma tissues compared with normal tissues. To confirm this result, 14 pairs of LUAD and noncancerous adjacent tissues were collected to examine the expression level of THAP7. Consistently, we found that THAP7 mRNA levels were significantly higher in LUAD tissues than in corresponding adjacent normal tissues (Figure 1B). We then measured the mRNA and protein levels of THAP7 in lung adenocarcinoma cell lines and the normal human bronchial epithelial cell line BEAS-2B. THAP7 showed increased expression in A549, H1299 and H1975 cell lines compared with normal BEAS-2B cells (Figure 1C and D). To evaluate whether the THAP7 mRNA level is clinically relevant, a large public clinical microarray database of lung adenocarcinoma was subjected to survival analysis. Importantly, we found that individuals with high THAP7 levels exhibited shorter overall survival (OS) (Figure 1E). Altogether, these results indicated that THAP7 might play a role in tumour promotion.
THAP7 promotes tumour growth in vitro and in vivo in lung adenocarcinoma
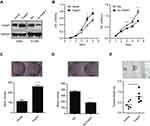
To further explore the biological function of THAP7 in lung adenocarcinoma cells, we overexpressed THAP7 in A549 cells and knocked down THAP7 in H1299 cells (Figure 2A). The results showed that the overexpression of THAP7 significantly promoted cell proliferation and colony formation (Figure 2B and C). In contrast, THAP7 silencing decreased H1299 cell growth and colony formation (Figure 2B and D). To verify the role of THAP7 on lung cancer growth in vivo, THAP7 overexpressing A549 cells and control cells were subcutaneously injected into nude mice to establish tumour xenograft. As shown in Figure 2E, the THAP7 overexpression group showed increased tumour growth compared to the control group.
THAP7 promotes the G1/S phase transition
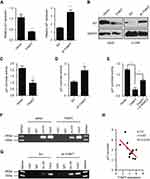
To elucidate the mechanism underlying the tumour promotion effect of THAP7 on LUAD, the cell cycle was analysed by flow cytometry. The results showed that THAP7 overexpression decreased the proportion of G1 cells and increased the proportion of S and G2 cells, while THAP7 silencing caused cell cycle arrest in G1 phase (Figure 3A and B). Taken together, these data suggest that THAP7 facilitates cell proliferation via acceleration of the G1/S phase transition.
THAP7 reduced the acetylation of histone H3 on the p21 promoter to inhibit p21 expression
P21 is a key regulator of the cell cycle in human cancer cells.26,27 We thus assessed whether THAP7 promoted cell cycle progression by regulating p21. The results of qRT-PCR and Western blotting revealed that the overexpression of THAP7 decreased both mRNA and protein levels of p21, which, in contrast, increased when THAP7 was silenced (Figure 4A and B). Given that p21 is an exceedingly regulated gene and its promoter activity is affected by various pathways,27 p21 promoter activity was estimated. The dual-luciferase reporter assay revealed that the ectopic expression or knockdown of THAP7 significantly decreased or increased the activity of the p21 promoter (Figure 4C and D), suggesting a key role for THAP7 in regulating p21 transcription in LUAD cells. THAP7 has been show to repress transcription by recruiting HDAC3.19–21 Thus, we performed rescue experiments. As shown in Figure 4E, knocking down HDAC3 by siRNA partly inhibited the THAP7-mediated suppression of p21 expression, suggesting that that THAP7-mediated suppression of p21 expression partly depends on HDAC3. Furthermore, ChIP assays showed that the overexpression or knockdown of THAP7 decreased or increased histone H3 acetylation in the p21 promoter region (Figure 4F and G). Moreover, we detected THAP7 and p21 expression at the mRNA levels and found a negative correlation between THAP7 and p21 in lung cancer tissues (Figure 4H). Collectively, these results suggest that THAP7 may inactivate p21 transcriptional activity via decreased H3 acetylation in the p21 promoter region.
Discussion
Although the advent of targeted therapies improved the outcomes in a subset of lung adenocarcinoma patients, most patients with metastatic adenocarcinoma received empirical chemotherapy and showed comparatively poor response rates.1,28 Therefore, it is of great importance to understand the molecular mechanisms of lung adenocarcinoma pathogenesis and identify novel therapeutic targets. In the present study, we showed that THAP7 is upregulated in lung adenocarcinoma and associated with worse clinical outcomes. By in vitro and in vivo studies, we revealed that THAP7 promoted tumour cell growth. We, for the first time, provided the evidence that THAP7 might be a promising biomarker and target for lung adenocarcinoma treatment.
A dysregulated cell cycle is often linked to increased tumourigenesis and accelerated tumour growth.29,30 The cell cycle contains cell growth (G1), DNA replication (S), and cell division (G2/M) phases. In general, the control of cell proliferation occurs at the G1 phase, during which the cells integrate multiple signals to determine entry into the S phase or exit to the G0 non-cycling quiescent phase.31 Most human cancers, including LUAD, are considered to develop from the disruption of G1/S cell cycle control.30 Interestingly, we also observed that THAP7 promoted G1/S cell cycle progression and cellular proliferation. A similar observation was obtained for another member of the THAP family, THAP1, which showed the regulation of endothelial cell proliferation through the modulation of pRB/E2F cell cycle target genes.12 In addition, the G1/S transition is controlled by cyclin/cyclin-dependent kinases (CDKs) and associated cyclin-dependent kinase inhibitors (CKIs). p21, a CKI, is a well-known tumour suppressor that regulates cell proliferation, and the dysregulation of p21 is a common feature across different cancer types, including LUAD.32,33 Here, we found that THAP7 reduced p21 transcription, enriching our knowledge of p21 expression regulation.
The epigenetic regulation of gene expression plays a crucial role in the progression of cancers, including lung cancer,34 and histone acetylation plays a key role in gene transcriptional regulation.35,36 Acetylation neutralizes the positive charge of histone lysine residues, producing a related chromatin conformation that allows access of the transcription machinery. In contrast, histone deacetylases (HDACs) could mediate histone deacetylation, which induces a condensed chromatin state and leads to transcriptional repression.37 THAP7 is a chromatin-associated protein, and the THAP domain is considered to exert transcriptional repression by binding to histones and nucleosomes in a tail-dependent manner and associating with chromatin. To explore the molecular mechanisms of the THAP7-mediated suppression of p21 expression, ChIP assays were performed using the anti-H3ac antibody, and the results suggested that THAP7 regulates p21 gene expression by reducing the acetylation of histone H3 on the p21 promoter, which might explain the inhibitory effect of THAP7 on p21 expression in LUAD. However, we still have not found the detailed mechanism by which the THAP7-mediated inhibition of p21 expression depends on other proteins besides HDAC3. These findings imply that THAP7 coordinates p21 signalling via multiple mechanisms.
Conclusion
The present study reports for the first time that THAP7 is upregulated in LUAD tissues and cell lines and is correlated with poor prognosis. Our findings highlight the importance of THAP7 in LUAD cell proliferation and provide evidence that the THAP7-p21 axis might serve as a potential valuable target for treating patients with LUAD.
Acknowledgments
This study was supported by the National Science Foundation of China (31501182, 81660449, and 81660414), the Jiangxi Provincial Natural Science Foundation of China (20161ACB21001 and 20171BCD40026), and the Jiangxi Provincial Health and Family Planning Commission Foundation (20164005 and 2015A077). We thank Jun Yang, Nanchang University, and Sheng-Jie Liu, China Pharmaceutical University, for technical assistance and manuscript revision. We thank Huai-Li Zhou, Xiaojing Wang and Jiaoping Mi, Sun Yat-sen University for technical assistance with the clinical sample collection and analysis.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi:10.1016/S0140-6736(16)30958-8
2. Nanavaty P, Alvarez MS, Alberts WM. Lung cancer screening: advantages, controversies, and applications. Cancer Control. 2014;21(1):9–14. doi:10.1177/107327481402100102
3. Sang Y, Sun L, Wu Y, Yuan W, Liu Y, Li SW. Histone deacetylase 7 inhibits plakoglobin expression to promote lung cancer cell growth and metastasis. Int J Oncol. 2019. doi:10.3892/ijo.2019.4682
4. Lv XB, Liu L, Cheng C, et al. SUN2 exerts tumor suppressor functions by suppressing the Warburg effect in lung cancer. Sci Rep. 2015;5:17940. doi:10.1038/srep17940
5. Feng LX, Wang J, Yu Z, et al. Clinical significance of serum EGFR gene mutation and serum tumor markers in predicting tyrosine kinase inhibitor efficacy in lung adenocarcinoma. Clin Transl Oncol. 2019. doi:10.1007/s12094-018-02014-6
6. Cai D, Choi PS, Gelbard M, Meyerson M. Identification and characterization of oncogenic SOS1 mutations in lung adenocarcinoma. Mol Cancer Res. 2019. doi:10.1158/1541-7786.MCR-18-0316
7. Xie L, Chen Y, Chen J, et al. Anti-tumor effects and mechanism of GA-13315, a novel gibberellin derivative, in human lung adenocarcinoma: an in vitro and in vivo study. Cell Mol Biol Lett. 2019;24:6. doi:10.1186/s11658-018-0126-9
8. Roussigne M, Cayrol C, Clouaire T, Amalric F, Girard JP. THAP1 is a nuclear proapoptotic factor that links prostate-apoptosis-response-4 (Par-4) to PML nuclear bodies. Oncogene. 2003;22(16):2432–2442. doi:10.1016/S0968-0004(02)00013-0
9. Roussigne M, Kossida S, Lavigne AC, et al. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci. 2003;28(2):66–69. doi:10.1016/S0968-0004(02)00013-0
10. Clouaire T, Roussigne M, Ecochard V, Mathe C, Amalric F, Girard JP. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc Natl Acad Sci U S A. 2005;102(19):6907–6912. doi:10.1073/pnas.0406882102
11. Gervais V, Campagne S, Durand J, Muller I, Milon A. NMR studies of a new family of DNA binding proteins: the THAP proteins. J Biomol NMR. 2013;56(1):3–15. doi:10.1007/s10858-012-9699-1
12. Cayrol C, Lacroix C, Mathe C, et al. The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood. 2007;109(2):584–594. doi:10.1182/blood-2006-03-012013
13. Poche RA, Zhang M, Rueda EM, et al. RONIN is an essential transcriptional regulator of genes required for mitochondrial function in the developing retina. Cell Rep. 2016;14(7):1684–1697. doi:10.1016/j.celrep.2016.01.039
14. Dejosez M, Krumenacker JS, Zitur LJ, et al. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133(7):1162–1174. doi:10.1016/j.cell.2008.05.047
15. Balakrishnan MP, Cilenti L, Mashak Z, Popat P, Alnemri ES, Zervos AS. THAP5 is a human cardiac-specific inhibitor of cell cycle that is cleaved by the proapoptotic Omi/HtrA2 protease during cell death. Am J Physiol Heart Circulatory Physiol. 2009;297(2):H643–653. doi:10.1152/ajpheart.00234.2009
16. Parker JB, Palchaudhuri S, Yin H, Wei J, Chakravarti D. A transcriptional regulatory role of the THAP11-HCF-1 complex in colon cancer cell function. Mol Cell Biol. 2012;32(9):1654–1670. doi:10.1128/MCB.06033-11
17. De Souza Santos E, De Bessa SA, Netto MM, Nagai MA. Silencing of LRRC49 and THAP10 genes by bidirectional promoter hypermethylation is a frequent event in breast cancer. Int J Oncol. 2008;33(1):25–31.
18. Lian WX, Yin RH, Kong XZ, et al. THAP11, a novel binding protein of PCBP1, negatively regulates CD44 alternative splicing and cell invasion in a human hepatoma cell line. FEBS Lett. 2012;586(10):1431–1438. doi:10.1016/j.febslet.2012.04.016
19. Macfarlan T, Kutney S, Altman B, Montross R, Yu J, Chakravarti D. Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor. J Biol Chem. 2005;280(8):7346–7358. doi:10.1074/jbc.M411675200
20. Macfarlan T, Parker JB, Nagata K, Chakravarti D. Thanatos-associated protein 7 associates with template activating factor-Ibeta and inhibits histone acetylation to repress transcription. Mol Endocrinol. 2006;20(2):335–347. doi:10.1210/me.2005-0248
21. Miele A, Medina R, van Wijnen AJ, Stein GS, Stein JL. The interactome of the histone gene regulatory factor HiNF-P suggests novel cell cycle related roles in transcriptional control and RNA processing. J Cell Biochem. 2007;102(1):136–148. doi:10.1002/jcb.21284
22. Chen CP, Chen X, Qiao YN, et al. In vivo roles for myosin phosphatase targeting subunit-1 phosphorylation sites T694 and T852 in bladder smooth muscle contraction. J Physiol. 2015;593(3):681–700. doi:10.1113/jphysiol.2014.283853
23. Chen CP, Chen K, Feng Z, Wen X, Sun H. Synergistic antitumor activity of artesunate and HDAC inhibitors through elevating heme synthesis via synergistic upregulation of ALAS1 expression. Acta Pharm Sin B. 2019. doi:10.1016/j.apsb.2019.05.001
24. Sang Y, Chen MY, Luo D, et al. TEL2 suppresses metastasis by down-regulating SERPINE1 in nasopharyngeal carcinoma. Oncotarget. 2015;6(30):29240–29253. doi:10.18632/oncotarget.5074
25. Sang Y, Cheng C, Zeng YX, Kang T. Snail promotes metastasis of nasopharyngeal carcinoma partly by down-regulating TEL2. Cancer Commun. 2018;38(1):58. doi:10.1186/s40880-018-0328-6
26. Sang Y, Zhang R, Sun L, et al. MORF4L1 suppresses cell proliferation, migration and invasion by increasing p21 and E-cadherin expression in nasopharyngeal carcinoma. Oncol Lett. 2019;17(1):294–302. doi:10.3892/ol.2018.9588
27. El-Deiry WS. p21(WAF1) mediates cell-cycle inhibition, relevant to cancer suppression and therapy. Cancer Res. 2016;76(18):5189–5191. doi:10.1158/0008-5472.CAN-16-2055
28. Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol. 2015;16(7):e342–351. doi:10.1016/S1470-2045(15)00077-7
29. Fu L, Kettner NM. The circadian clock in cancer development and therapy. Prog Mol Biol Transl Sci. 2013;119:221–282. doi:10.1016/B978-0-12-396971-2.00009-9
30. Lee Y, Lahens NF, Zhang S, Bedont J, Field JM, Sehgal A. G1/S cell cycle regulators mediate effects of circadian dysregulation on tumor growth and provide targets for timed anticancer treatment. PLoS Biol. 2019;17(4):e3000228. doi:10.1371/journal.pbio.3000171
31. Duronio RJ, Xiong Y. Signaling pathways that control cell proliferation. Cold Spring Harb Perspect Biol. 2013;5(3):a008904. doi:10.1101/cshperspect.a008904
32. Zhu L, Wang J, Kong W, et al. LSD1 inhibition suppresses the growth of clear cell renal cell carcinoma via upregulating P21 signaling. Acta Pharmaceutica Sinica B. 2019;9(2):324–334. doi:10.1016/j.apsb.2018.10.006
33. Luo J, Liu K, Yao Y, et al. DMBX1 promotes tumor proliferation and regulates cell cycle progression via repressing OTX2-mediated transcription of p21 in lung adenocarcinoma cell. Cancer Lett. 2019;453:45–56. doi:10.1016/j.canlet.2019.03.045
34. Zhao L, Duan YT, Wang JL, et al. Epigenetic targets and their inhibitors in cancer therapy. Curr Top Med Chem. 2018;18(28):2395–2419. doi:10.2174/1568026619666181224095449.
35. Clocchiatti A, Florean C, Brancolini C. Class IIa HDACs: from important roles in differentiation to possible implications in tumourigenesis. J Cell Mol Med. 2011;15(9):1833–1846. doi:10.1111/j.1582-4934.2011.01321.x
36. Guerriero JL, Sotayo A, Ponichtera HE, et al. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature. 2017;543(7645):428–432. doi:10.1038/nature21409
37. Lasko LM, Jakob CG, Edalji RP, et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature. 2017;550(7674):128–132. doi:10.1038/nature24028
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.