Back to Journals » Cancer Management and Research » Volume 12
TCEAL2 as a Tumor Suppressor in Renal Cell Carcinoma is Associated with the Good Prognosis of Patients
Authors Zhou Y , Zhang Y, Li W, Xu J, He X, Li X, Wang Y
Received 17 July 2020
Accepted for publication 4 September 2020
Published 2 October 2020 Volume 2020:12 Pages 9589—9597
DOI https://doi.org/10.2147/CMAR.S271647
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Yingchen Zhou,1,2 Yang Zhang,2 Weiqing Li,2 Jinming Xu,2 Xia He,1 Xianxin Li,1– 3 Yan Wang2
1Department of Surgery, Fuwai Hospital Chinese Academic of Medical Science Shenzhen, The University of South China, Shenzhen, People’s Republic of China; 2Department of Urology, Peking University Shenzhen Hospital, Shenzhen PKU-HKUST Medical Center, Shenzhen, People’s Republic of China; 3Department of Urology, Taikang Qianhai International Hospital, Shenzhen, People’s Republic of China
Correspondence: Xianxin Li
Department of Urology, Peking University Shenzhen Hospital, Lianhua Road 1120, Shenzhen 518036, People’s Republic of China
Tel +86-755-83923333-3419
Fax +86 755-83910721
Email [email protected]
Yan Wang
Department of Urology, Peking University Shenzhen Hospital, Lianhua Road 1120, Shenzhen 518036, People’s Republic of China
Tel +86-755-83923333-3419
Fax +86 755-83910721
Email [email protected]
Background: Renal cell carcinoma (RCC) is one of the most common tumors in urinary tract tumors. However, the mechanism that supports renal cell carcinoma is unclear. The function of transcription elongation factor A (SII)-like 2 (TCEAL2) and its association with human cancer have not been reported.
Materials and Methods: To explore the role of TCEAL2 in carcinogenesis of clear cell renal cell carcinoma (ccRCC), we performed bioinformatics analysis to determine the expression levels of TCEAL2 in ccRCC specimens and normal kidney tissue and then verified findings with our samples by qPCR, Western blot and immunohistochemistry staining. Furthermore, the lentiviral vectors were used to increase the expression of TCEAL2 in ccRCC cell lines. The immunofluorescence assay was taken to observe the subcellular location of TCEAL2 in ccRCC cells, and CCK-8 and flow cytometry were introduced for assessing cell proliferation and cell cycle of ccRCC cells, respectively.
Results: Compared with adjacent normal kidney tissue and human proximal tubular epithelial cells, the expression of TCEAL2 in ccRCC tissues and cell lines was down-regulated. Patients who had low expression of TCEAL2 had a statistically significant late tumor stage. Restore of TCEAL2 in ccRCC cells inhibited cell proliferation and induced cell cycle arrest in S phase of ccRCC cells.
Conclusion: To our knowledge, this is the first report of TCEAL2 expression changes in ccRCC. We found that the decrease of TCEAL2 expression may be related to the occurrence of ccRCC. Further research is needed to clarify the molecular mechanism of TCEAL2 in progress of ccRCC.
Keywords: TCEAL2, renal cell carcinoma, tumorigenesis, cell cycle
Introduction
Renal cell carcinoma (RCC), also known as renal adenocarcinoma, is one of the common tumors in the urinary system and the second leading cause of death in the world. About 200,000 people were diagnosed with renal cell carcinoma every year around worldwide. The national age-standardized incidence rate is 12/100,000, accounting for 2% to 3% of adult malignancies, and accounting for approximately 90% of kidney tumors, and its incidence is increasing in Europe.1 Renal cell carcinoma originates from renal tubular epithelial cells, clear cell carcinoma of the kidney is most common.2 Its treatment plan is still based on radical surgery. Postoperative adjuvant treatment options are diverse, but effective adjuvant treatment options are lacking.3 However, due to asymptomatic and non-specific markers in the early stages of renal cancer, most patients come to the hospital when tumor immersion and painless hematuria appear in the renal pelvis.4 Therefore, an exploration of potential molecular mechanisms and the identification of effective biomarkers involved in the pathogenesis of clear cell renal cell carcinoma (ccRCC) is urgently needed to improve the effectiveness of therapeutic strategies.
The transcription elongation factor A-like 2 (TCEAL2) gene (also known as WEX1, my048 and MY0876G05) was first cloned and located on the Xq22.1 chromosome in 2005.5,6 It spans 2025 bp and consists of three exons. Human TCEAL2 mRNA is 1100 bp long and encodes a protein of 227 amino acids with a relative molecular mass of approximately 26 kDa.7 TCEAL2 belongs to the transcription elongation factor A (SII)-like (TCEAL) gene family containing TFA domains.8 It has been reported as a nuclear phosphoprotein that modulates transcription in a promoter context-dependent manner and has been recognized as an important nuclear target for intracellular signal transduction.8 It was reported that up-regulation of TCEAL2 might be associated with a poor prognosis patients with ovarian cancer.9 Recent studies have shown that TCEAL2 expression was found to be reduced in ccRCC, TCGC and TCC whole-genome sequencing.10 However, the expression and physiological effects of TCEAL2 in ccRCC have not been studied so far.
Here, we confirmed that TCEAL2 is generally reduced in the mRNA and protein levels of ccRCC tissues and cell lines, and the decreased TCEAL2 expression is related to the clinicopathological parameters of renal cell carcinoma. Therefore, the change of TCEAL2 protein level might be used for early detection of RCC, monitoring tumor progression and predicting clinical markers of patient prognosis.
Materials and Methods
Cell Culture and ccRCC Tissue Specimens
The clear cell renal cell carcinoma cells (786-O, 769-P, ACHN, Caki-1 and Caki-2) and human proximal tubular epithelial cell line (HK-2) were obtained from the American Type Culture Collection. All cell lines were maintained in culture mediums as described previously.11 All tissue samples for RT-qPCR were obtained from ccRCC patients without undergoing radiotherapy and chemotherapy between 2013 and 2015 at the Department of Urology, Peking University Shenzhen Hospital, China. All experiments followed the “Helsinki Declaration” and were approved by the Ethics Committee of Peking University Shenzhen Hospital (NO.20090017). All patients were informed of their specimen content, potential risks, purpose and signed written informed consent.
Database Mining
The TCGA data and GEO dataset GSE53757 were downloaded to investigate the expression of TCEAL2 mRNA levels in ccRCC and normal kidney tissues, and the correlation of TCEAL2 and clinicopathological characteristics of patients with ccRCC on April 18, 2019.
RT-qPCR
Total RNA was isolated from freshly frozen clear cell renal cell carcinoma tissues or cell lines using RNAiso Plus lysis reagent (Takara, Japan). One microgram of total RNA was reversely transcribed to cDNA using the PrimeScript RT reagent Kit (Takara, Japan). The TCEAL2 gene primer amplification sequence: forward: 5ʹ-AACATGGCTAGGGTGGAGGA-3ʹ; reverse: 5ʹ-TCCACGAATCATGACGTCTCT-3ʹ. The human GAPDH gene was used as an endogenous control for RNA standardization and amplified with the following primer pairs: forward: 5ʹ-GCTCTCCAGAACATCATCCCTGCC-3ʹ; reverse: 5ʹ-CGTTGTCATACCAGGAAATGAGCTT-3ʹ. The qPCR was performed using SYBR Premix Ex TaqTMII Kit (Takara, Japan).
Western Blotting
Western blotting was performed as described previously.12 Primary antibodies used were against TCEAL2 (1:1000, Santa Cruz Biotechnology, Inc. USA) and β-tubulin (1:5000, Abcam, UK). The secondary antibody was anti-rabbit (1:2000, Cell Signaling Technology, USA). Luminescence of protein bands was observed by using a chemiluminescence imaging system (Tanon-5200Multi, Tanon, China).
Immunohistochemistry Staining
Immunohistochemistry staining was performed on Single spot tissue micro arrays (TMA) slides (90 points of RCC, 90 points of adjacent normal tissue, HKid-CRC180Sur-01, Shaanxi Avila Biotechnology Co., Ltd, Shaanxi, China) using the TCEAL2 rabbit antibody (1:100, Santa Cruz Biotechnology, Inc. USA) and the rabbit streptavidin-biotin detection system (Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd. Peking, China).
The TMA was manually scored by two pathologists (X. H. and G.Y.) according to a 4-layer scoring system. We defined tumor cells with ≥30% strong staining or ≥70% moderate staining as a score of 3+. Tumor cells with >70% weak staining or >30 and ≤70% medium staining or ≤30% strong tumor cell staining were defined as a score of 2+. When ≤70% of tumor cells were weakly positive or ≤30% were moderately stained, the score was 1+. Less staining was defined as negative (score 0). Inconsistent results are resolved through consensus review.
Immunofluorescence Staining
For immunofluorescence, 1 x 10^4 ccRCC cells with or without TCEAL2 overexpression were seeded on cell slides. Cells in logarithmic phase were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with blocking buffer, and incubated with primary antibody against TCEAL2 overnight at 4°C. Samples were then labeled with fluorochrome-conjugated secondary antibodies (Life Technology, USA). Finally, DAPI staining for DNA was performed. Cells covered with fluorescence mounting medium (DAKO, CA, USA) were examined using a fluorescence system mounted on a BX53 upright microscope (Olympus, Japan).
Establishment of ccRCC Cells with TCEAL2 Stable Expression
The lentiviral vector GV358-TCEAL2 and control vector were obtained from Genechem technology company (Shanghai, China). To establish ccRCC cells stably expressing TCEAL2, 2 x 10^4 ccRCC cells per well were seeded in 6-well plate and infected with lentiviral vector GV358-TCEAL2. One-milliliter fresh culture mediums were added in cells at 24 h after infection. The cells were observed under fluorescent microscope at 72 h after infection.
Cell Proliferation
3 x 10^3/well ccRCC cells with lentiviral vector GV358-TCEAL2 or control vector were inoculated into 96-well plates, and the proliferation was analyzed at 0, 24, 48 and 72 hours by using cell counting kit 8 (CCK-8) (Medium: Cell counting kit 8=10:1, Dojindo Laboratories, Kumamoto, Japan).
Flow Cytometry for Cell Cycle
ccRCC cells with or without TCEAL2 stable expression cultured for 48h were collected and washed twice with PBS, and then fixed with 70% ice-cold ethanol overnight at 4°C. The fixed cells were resuspended in PBS containing PI/RNase staining buffer (BD Biosciences, San Diego, CA), and incubated for 15 minutes at room temperature. Cells were subjected to flow cytometric analysis of DNA content using a flow cytometer (BD AccuriTM C6 Plus, BD Bioscience, CA). The percentages of cell cycle distribution were calculated by Flowjo VX software.
Statistical Analysis
All data from three repeated experimental results are expressed as mean ± SD. The correlation of TCEAL2 and each clinical pathological variable was comparatively analyzed by χ2 test, the Fisher exact test and Chi-square test. A p value of less than 0.05 indicated the presence of statistically significant difference between groups. All statistical analyses were carried out with SPASS 19.0 and GraphPad Prism 6 software.
Results
TCEAL2 Was Down-Regulated in ccRCC Tissues and Cell Lines
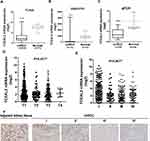
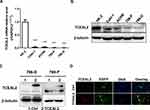
To identify our preliminary findings, the TCGA visual database GEPIA and cBioPortal and the GEO Profile GSE53757 were adopted. The expression of TCEAL2 in patients with ccRCC was significantly lower than that in patients without ccRCC (Figure 1A and B). In addition, immunohistochemical staining showed that 82 cases (91.1%) of RCC cases had negative expression of TCEAL2 in cancer tissues, while only 8 cases (8.9%) of adjacent normal tissues had low expression of TCEAL2 (P <0.001) (Table 1). By analyzing the clinicopathological data of 90 patients with tissue microarray, the expression of TCEAL2 was not related to the patient’s tumor stage, and the difference was not statistically significant (P>0.05) (Table 2). Furthermore, in accordance with the immunohistochemical staining, RT-qPCR data also showed a significant decrease of TCEAL2 mRNA in 20 cases of ccRCC tissues compared to the adjacent normal kidney tissues (P<0.001) (Figure 1C and F). And in the TCGA visual database, the expression of TCEAL2 was related to the tumor stage and T grade in renal cancer patients (P<0.001) (Figure 1D and E). Then, TCEAL2 expression levels were detected in ccRCC cell lines (786-O, 769-P, ACHN and Caki-1) and normal renal tubular epithelial cell lines (HK-2). As shown in Figure 2A and B, TCEAL2 expression was significantly reduced in the 786-O, 769-P, ACHN and Caki-1 cell lines at both transcription and translation levels, compared to the HK-2 cell line (p<0.001, respectively).
 |
Table 1 Expression of TCEAL2 in ccRCC and Adjacent Normal Kidney Tissues |
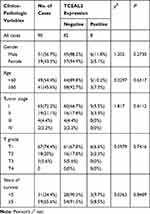 |
Table 2 Correlation Between TCEAL2 Expression and the Clinicopathological Characters of Patients with Clear Cell Renal Cell Carcinoma (IHC) |
TCEAL2 Under-Expression is Associated with the Tumor Stage of Patients with ccRCC
We analyzed the possible correlation between the TCEAL2 mRNA level in 534 ccRCC samples and the clinical characteristics of ccRCC through TCGA data. As summarized in Table 3, patients were divided into two subgroups based on TCEAL2 expression level: low-expression group (n = 510) and high-expression group (n = 24). Compared with the high-expression group, the TCEAL2 low-expression group more appeared in advanced tumor stage (P = 0.0364), T grade (P = 0.0466) and male (P = 0.0073). There was no significant correlation between the expression level of TCEAL2 and age and 5-year survival rate (Table 3). According to the above-mentioned TCEAL2 mRNA level and clinicopathological characteristics of ccRCC patients, the multivariate cox-regression analysis showed that the patient’s age and tumor stage were related with survival time, and the risk coefficients are 1.428 and 1.988, respectively (Table 4). However, the expression of TCEAL2 in ccRCC has not been a variable that affects their survival time, which may require more samples for evaluation.
 |
Table 3 Correlation Between TCEAL2 Expression and the Clinicopathological Characters of Patients with Clear Cell Renal Cell Carcinoma (TCGA) |
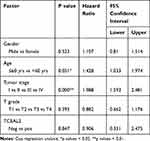 |
Table 4 Multivariate Cox-Regression Analysis for Patients After Surgery (TCGA) |
TCEAL2 Can Inhibit the Proliferation of ccRCC Cells
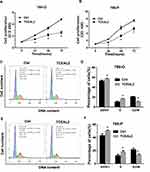
To further identify the biological significance of TCEAL2 in ccRCC cells, we first performed the lentivirus to overexpress TCEAL2. As shown in results (Figure 2C), after TCEAL2 lentivirus infection, the protein level of TCEAL2 in ccRCC cell lines 786-O and 769-P was significantly higher than that in the control group by Western blot. It indicated that we successfully overexpressed TCEAL2 using lentiviral vector. Immunofluorescence data showed that TCEAL2 was located in the nucleus (Figure 2D). CCK-8 assay showed that ccRCC cell lines 786-O and 769-P with TCEAL2 overexpression had notable decreased proliferation ability compared to the control group (Figure 3A-B). This indicated that TCEAL2 might be associated with tumorigenesis of ccRCC.
TCEAL2 Induces Cell Cycle Arrest in S Phase of ccRCC Cells
To better understand why cells overexpressing TCEAL2 proliferated slowly, the cell population of ccRCC cells with or without TCEAL2 stable expression was analyzed by flow cytometry, and the role of TCEAL2 on cell cycle progression was evaluated. As shown in Figure 3, a significantly lower G2/M phase percentage (15.1% for 786-O and 10.1% for 769-P) was observed in TCEAL2 stably expressing ccRCC cells than that in control group cells (22.1% for 786-O and 20.8% for 769-P), while a significantly higher G0/G1 (51.0% for 786-O and 56.2% for 769-P) or S phase (16.2% for 786-O and 15.9% for 769-P) percentage was observed in ccRCC cells stably expressing TCEAL2 than that in control group cells. The above results indicated that TCEAL2 is correlated with the cell cycle progression in ccRCC.
Discussion
The elongation of transcription in eukaryotes is a very complicated process.13 When RNAPII slides along the DNA template, it will encounter obstacles and produce instantaneous pauses and arrests. Some protein factors can directly or indirectly interact with RNAP II during this process to inhibit or affect its transient rest and stagnation and thus affect the efficiency of transcription. A handful of studies showed that transcription elongation factors involved in human ccRCC tumorigenesis and metastasis. Two transcription elongation factors, Elongin B and C were shown to bind in vitro and in vivo to a short, colinear region of the VHL protein (pVHL) that is frequently mutated in human ccRCC.14–16 In our previous study, we found that transcription elongation factor A-like protein TCEAL2 was down-regulated in ccRCC tissues.10 In this study, we firstly investigated the expression and function of TCEAL2 in clear cell renal cell carcinoma. Compared with normal kidney tissue, the expression of TCEAL2 in ccRCC was significantly reduced at mRNA and protein levels. In addition, the low expression of TCEAL2 was associated with a higher tumor stage and male.
TCEAL2 like other members of the transcription elongation factor A-like family contains a BEX protein conserved domain that regulates gene transcription. Tceal1/SIIR/p21, a nuclear phosphoprotein, can inhibit the transcriptional activity of the long end of Rous sarcoma virus.8,17 Down-regulation of Tceal7 encoding a cell death-regulating protein18 enhances the transcriptional activity of NF-kB by increasing the expression levels of the cell-promoting genes cyclin D1 and c-Myc, and the angiogenic genes IL-6, IL-8, and VEGF.19 Up to date, there are few studies about the expression of TCEAL2 in cancers. Kim et al have found up-regulation of TCEAL2 might be associated with a poor prognosis of stage III serous ovarian cancer.9 Here, we reported that under-expression of TCEAL2 in ccRCC was associated with higher tumor stage, and TCEAL2 could inhibit cell proliferation and induced cell cycle arrest into S phase.
In addition to the BEX protein domain, TCEAL2 also contains a K+-dependent Na+/Ca+ exchanger (data not shown). It is reported that K+-dependent Na+/Ca+ exchanger can inhibit the proliferation of tumor cells and induce apoptosis by regulating the concentration of calcium ions inside and outside the cell.20–22 Since TCEAL2 contains the conserved domains BEX and K+-dependent Na+/Ca+ exchanger, we speculate that TCEAL2 may have similar functions with other members of the transcription elongation factor A-like family.
In short, this is the first study to describe the role of TCEAL2 expression in the development of renal clear cell carcinoma. The results suggest that loss of expression of TCEAL2 might play a role as a tumor suppressor in clear cell renal cell carcinoma. To clarify the mechanisms of TCEAL2 in renal cancer, further molecular biology experiments are required.
Abbreviations
RCC, renal cell carcinoma; TCEAL2, transcription elongation factor A (SII)-like 2; ccRCC, clear cell renal cell carcinoma.
Funding
This study was supported by the grant from the Science Technology and Innovation Commission of Shenzhen Municipality (NO. JCYJ20170307161640543 and NO. JCYJ20190809104411245).
Disclosure
The authors have no conflicts of interest to declare for this work.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi:10.1002/ijc.29210
2. Klaassen Z, Sayyid RK, Wallis CJD. Lessons learned from the global epidemiology of kidney cancer: a refresher in epidemiology 101. Eur Urol. 2019;75(1):85–87. doi:10.1016/j.eururo.2018.09.035
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
4. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
5. Ross MT, Grafham DV, Coffey AJ, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337.
6. Winter EE, Ponting CP. Mammalian BEX, WEX and GASP genes: coding and non-coding chimaerism sustained by gene conversion events. BMC Evol Biol. 2005;5(1):54. doi:10.1186/1471-2148-5-54
7. Büssow K, Cahill D, Nietfeld W, et al. A method for global protein expression and antibody screening on high-density filters of an arrayed cDNA library. Nucleic Acids Res. 1998;26(21):5007–5008. doi:10.1093/nar/26.21.5007
8. Pillutla RC, Shimamoto A, Furuichi Y, et al. Genomic structure and chromosomal localization of TCEAL1, a human gene encoding the nuclear phosphoprotein p21/SIIR. Genomics. 1999;56(2):217–220. doi:10.1006/geno.1998.5705
9. Kim Y-S, Hwan JD, Bae S, et al. Identification of differentially expressed genes using an annealing control primer system in stage III serous ovarian carcinoma. BMC Cancer. 2010;10:576. doi:10.1186/1471-2407-10-576
10. Li X, Chen J, Hu X, et al. Comparative mRNA and microRNA expression profiling of three genitourinary cancers reveals common hallmarks and cancer-specific molecular events. PLoS One. 2011;6(7):e22570. doi:10.1371/journal.pone.0022570
11. Wang Y, Dong D, Jiang S, et al. miR-216b post-transcriptionally downregulates oncogene KRAS and inhibits cell proliferation and invasion in clear cell renal cell carcinoma. Cell Physiol Biochem. 2018;49(5):1755–1765. doi:10.1159/000493621
12. Wang Y, Zhao Y, Herbst A, et al. miR-221 mediates chemoresistance of esophageal adenocarcinoma by direct targeting of DKK2 expression. Ann Surg. 2016;264(5):804–814. doi:10.1097/SLA.0000000000001928
13. Selth LA, Sigurdsson S, Svejstrup JQ. Transcript elongation by RNA polymerase II. Annu Rev Biochem. 2010;79(1):271–293. doi:10.1146/annurev.biochem.78.062807.091425
14. Ohh M, Kaelin WG. VHL and kidney cancer. Methods Mol Biol. 2003;222:167–183.
15. Maher ER, Kaelin WG. von Hippel-Lindau disease. Medicine (Baltimore). 1997;76(6):381–391. doi:10.1097/00005792-199711000-00001
16. Kibel A, Iliopoulos O, DeCaprio JA, et al. Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science. 1995;269(5229):1444–1446. doi:10.1126/science.7660130
17. Yeh CH, Shatkin AJ. A cis-acting element in Rous sarcoma virus long terminal repeat required for promoter repression by heLa nuclear protein p21. J Biol Chem. 1995;270(26):15815–15820. doi:10.1074/jbc.270.26.15815
18. Chien J, Staub J, Avula R, et al. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24(32):5089–5100. doi:10.1038/sj.onc.1208700
19. Rattan R, Narita K, Chien J, et al. TCEAL7, a putative tumor suppressor gene, negatively regulates NF-kappaB pathway. Oncogene. 2010;29:1362–1373.
20. Gylfe AE, Kondelin J, Turunen M, et al. Identification of candidate oncogenes in human colorectal cancers with microsatellite instability. Gastroenterology. 2013;145:540–543.e22. doi:10.1053/j.gastro.2013.05.015
21. Zhang Z, Sun S, Du C, et al. Effects of leptin on Na+/Ca2+ exchanger in PC12 cells. Cell Physiol Biochem. 2016;40(6):1529–1537. doi:10.1159/000453203
22. Xu J, Ji B, Wen G, et al. Na+/Ca2+ exchanger 1 and calmodulin complex regulates interleukin 6-mediated cellular behavior of human hepatocellular carcinoma. Carcinogenesis. 2016;37(3):290–300. doi:10.1093/carcin/bgw004
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.