Back to Journals » OncoTargets and Therapy » Volume 13
Targeting PIN-1 Attenuates GCB DLBCL Cell Proliferation Through Inhibition of PI3K/AKT Signaling
Authors Yang H, Zhang P, Li J, Gao Y , Zhao L, Li J, Guo M, Zhang J, Li H, Wang F, Yuan Y
Received 28 January 2020
Accepted for publication 26 July 2020
Published 25 August 2020 Volume 2020:13 Pages 8593—8600
DOI https://doi.org/10.2147/OTT.S247429
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Haijun Yang,1 Ping Zhang,1 Junkuo Li,1 Yang Gao,2 Luyao Zhao,2 Jia Li,1 Mei Guo,1 Jingfang Zhang,1 Haimei Li,1 Fuqiang Wang,1 Yufen Yuan1
1Department of Pathology, Anyang Tumor Hospital, The Fourth Affiliated Hospital of Henan University of Science and Technology, Anyang 455000, People’s Republic of China; 2NHC Key Laboratory of Biotechnology of Antibiotics, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100032, People’s Republic of China
Correspondence: Yufen Yuan Tel +86 18810845563
Email [email protected]
Introduction: Diffuse large B cell lymphoma (DLBCL) is a highly heterogeneous type of non-Hodgkin lymphoma with many molecular subtypes that can be distinguished by gene expression profiling (GEP). However, the pathogenesis of DLBCL is still unclear.
Materials and Methods: The expression levels of the prolyl isomerase PIN-1 and other related proteins were determined in 73 primary DLBCL patient samples and cell lines by Western blotting (WB) and immunohistochemical (IHC) staining. Cell cycle and apoptosis were evaluated by flow cytometry. Lymphoma cell viability was detected by CCK-8 proliferation assay.
Results: High levels of PIN-1 expression were detected in 55% of germinal center B cell (GCB) DLBCL patient samples, whereas such abnormal expression levels were found in only 11% of non-GCB DLBCL patient samples. PIN-1 expression was positively associated with activation of the oncogenic phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) pathway in both GCB DLBCL cell lines and primary patient samples. Depletion of PIN-1 was cytotoxic to GCB DLBCL model cell lines because it led to inhibition of the PI3K/AKT signaling pathway, revealing a GCB DLBCL subgroup that is dependent on this pathway. A PI3K inhibitor was selectively toxic to GCB DLBCL lines expressing high levels of PIN-1.
Conclusion: Our study used PIN-1 to identify a new subgroup of GCB DLBCL associated with the PI3K/AKT signaling pathway, and our findings reveal that inhibition of PI3K is a promising therapeutic approach for GCB DLBCL.
Keywords: PIN-1, DLBCL, PI3K/AKT signaling pathway
Introduction
Diffuse large B cell lymphoma (DLBCL), the most common subtype of non-Hodgkin lymphoma (NHL), accounts for almost one-third of all NHL cases.1 According to the unique genetic signatures identified by gene expression profiling (GEP) studies, DLBCL can itself be divided into three different molecular subtypes. The first is the germinal center B cell (GCB) subtype, which is characterized by gene expression levels typical of normal germinal B cells and has a good prognosis. The second subtype is the activated B cell (ABC)-like subtype, which is characterized by gene expression levels typical of activated peripheral blood B cells and plasma cells and has a poor prognosis. Other heterogeneous types without these characteristics are grouped into the third subtype, the prognosis of which is also poor.2 However, even within these defined subtypes, heterogeneity prevails.
The peptidyl-prolyl cis/trans isomerase PIN-1 can specifically recognize and bind phosphorylated serine or threonine residues preceding proline residues (p-Ser/Thr-Pro) and induce conformational changes in phosphoproteins.3 PIN-1 also transmits phosphorylation signals by affecting its substrate.4,5 PIN-1 alterations can stabilize cancerous proteins and/or inactivate or destabilize tumor suppressors, thus amplifying oncogenic signals, as has been suggested by the frequent deregulation of PIN-1 in several human malignancies.6 Moreover, recent research has revealed that PIN-1 plays a pivotal role in the progression of many cancers, such as T cell acute lymphocytic leukemia (T-ALL) and B cell acute lymphocytic leukemia (B-ALL).7,8 However, whether PIN-1 is involved in the pathogenesis of DLBCL and how PIN-1 regulates other oncogenic signaling pathways in DLBCL remain unknown.
The phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) signaling pathway plays crucial roles in multiple cellular processes. This pathway is often dysregulated in many cancers, including B cell lymphomas.9,10 PIN-1 can regulate the stability of the AKT protein and the phosphorylation of this protein at S473 by binding PIN-1 phosphorylated Thr-Pro residues at AKT T92 and T450 (the “turn motif”).11 PIN-1 deficiency or depletion compromises the ability of PIN-1 to protect AKT from degradation. Interestingly, abnormal PIN-1 expression levels are not detected in ABC DLBCL, suggesting that PIN-1 participates in the pathogenesis of only GCB DLBCL. We therefore hypothesized that PIN-1 may play a functional role in DLBCL. Hence, in this study we detected the expression of PIN-1 in the GCB and non-GCB subtypes of DLBCL for the first time. Furthermore, we identified that PIN-1 loss is cytotoxic to GCB DLBCL model cell lines because it leads to inhibition the PI3K/AKT signaling pathway. Overall, our study provides a rationale for PI3K inhibition as a promising therapeutic approach for GCB DLBCL.
Materials and Methods
Patient Samples
Primary cells were freshly obtained from the lymphoma tissues of lymphoma patients at Anyang Tumor Hospital, Henan University of Science and Technology. The clinical features of the patients are listed in Table S1. Informed consent was obtained from all the participants in accordance with the Declaration of Helsinki. Informed consent for any of the patients who were under the age of 18 was signed by a parent or legal guardian. All protocols using human specimens were approved by the Institutional Review Board of Anyang Tumor Hospital, Henan University of Science and Technology.
Cell Lines and LY294002 Treatment
BJAB is GCB DLBCL cell line and HBL-1 is non-GCB DLBCL cell line. They were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China), where they had recently been authenticated by short tandem repeat (STR) profiling and tested for mycoplasma contamination and cell vitality. The BJAB and HBL-1 cells were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) containing 10% heat-inactivated fetal bovine serum (FBS, Gibco) and 10 U/mL penicillin-streptomycin (Sigma, USA) in a humidified incubator at 37°C in 5% CO2 and 95% air. LY294002 was purchased from Sigma (USA), dissolved in 10% dimethyl sulfoxide (DMSO) and stored at −20°C. When DMSO was used as a vehicle control, the concentration was kept under 0.1% throughout all experiments to avoid cytotoxic effects.
Transfection
To stably express PIN-1 in BJAB and HBL-1 cells, Lipofectamine® 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) was used to transiently transfect the cells with control shRNA (shCTRL) or PIN-1 shRNA (PIN-1-shRNA-1/2) plasmids12 according to the manufacturer’s instructions. After 24 hours of transfection, 200 μg/mL hygromycin (Calbiochem, San Diego, CA, USA) was added to the medium, and the cells were cultured for 7 days for selection of stable transfectants. The stably transfected cells were screened for subsequent experiments through several passages in medium containing hygromycin.
Cell Growth and Apoptosis Analyses
Growth curves were determined in triplicate, and cell viability was quantified using a Vi-CELL XR Cell Viability Analyzer (Beckman Coulter, Brea, CA, USA). Apoptosis in lymphoma cells was analyzed with an apoptosis assay. Briefly, the cells were cultured in 6-well plates, collected, washed twice with phosphate-buffered saline (PBS), fixed in 75% cold ethanol overnight, and stained with annexin V-FITC/propidium iodide (PI) (GE Healthcare, PA, USA) in PBS for 30 min in the dark. The percentages of apoptotic cells were calculated by determining the proportions of sub-G1 cells using flow cytometry (GE Healthcare, PA, USA).
Western Blotting (WB)
In this study, the protein expression levels of PIN-1, AKT, p-AKT (T308) and other proteins were examined by WB. Cells were washed with ice-cold PBS (pH 7.4) and then centrifuged at 1000 rpm for 5 min. The cell pellets were lysed using RIPA lysis buffer (Beyotime, Jiangsu, China) with a protease inhibitor mixture (Selleck Chemicals, Houston, Texas, USA) and 1 mM PMSF. Polyvinylidene fluoride membranes to which PIN-1, AKT, p-AKT and other proteins had been transferred were incubated with primary antibodies (1:1000) and then with horseradish peroxidase-conjugated secondary antibodies (1:2000). The proteins were revealed using an enhanced chemiluminescence detection method following the manufacturer’s instructions (Pierce). Semiquantification of the positive bands in the images was performed using Quantity One software (Bio-Rad, CA, USA). The intensities of the experimental bands were normalized against the intensities of the bands for the internal control GAPDH minus the background.
Immunohistochemical Staining (IHC)
For IHC, deparaffinized and rehydrated slides were subjected to autoclave antigen retrieval in 10 mmol/L citric acid buffer (pH 6.0) and allowed to cool to room temperature. The slides were blocked with 3% H2O2 for 30 min, washed in PBS, and then blocked with 5% normal goat serum in PBS. The slides were incubated with diluted PIN-1 antibodies (Santa Cruz, sc-46660, 1:100) overnight at 4°C. PIN-1 staining was established by an expert hematopathologist. To differentiate PIN-1-positive and PIN-1-negative DLBCL cases, we applied the commonly used cutoff value of 30% positive cells. Using this cutoff, we were able to reproduce the expression levels detected by WB in DLBCL cell lines.
Cell‐of‐origin (COO) Groups
Based on the IHC results of three proteins, CD10, BCL6 and MUM1, the Hans algorithm was used to identify two subtypes of DLBCL: GCB and non‐GCB. In addition, a NanoString Technologies Lymph2Cx assay was used to classify DLBCL cases into two subgroups: ABC and non‐GCB. In total, the COO groups were successfully determined for 47 of 74 cases with the NanoString Lymph2Cx assay, whereas the analysis failed for 27 cases due to insufficient RNA quality. This information is presented in Supplementary Table S1.
Cell Cycle Analysis
Cells were cultured for 24 hours and then digested with 0.25% trypsin (without EDTA) into individual cells. The cells were collected by centrifugation, and the supernatant was discarded. The cells were washed twice with precooled PBS. Three milliliters of precooled 70% ethanol was added to the cell precipitate, and the cells were fixed overnight at 4°C. The cells were collected by centrifugation, washed twice with 3 mL of PBS, and then stained with PI and RNase A (Sigma). The cells were then incubated at 4°C for 30 min without light. PI fluorescence intensity was analyzed by a flow cytometer using the FL-2 channel.
Statistical Analysis
All data are presented as the means ± SEMs of three or more independent experiments. Statistical comparisons between two groups were performed using Student’s t-test. One-way ANOVA was used to analyze differences among more than two groups. GraphPad Software Prism 6.0 was used for statistical analysis. The statistically significant difference thresholds were set at *p < 0.05 and **p < 0.01.
Results
PIN-1 is Highly Expressed in GCB DLBCL
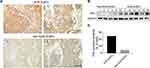
To explore the biological functions of PIN-1 in molecular DLBCL subtypes, we first determined the expression of PIN-1 in DLBCL patients. Seventy-three DLBCL patient samples were classified into the GCB DLBCL and non-GCB DLBCL subtypes by IHC, and the expression of PIN-1 was further examined by IHC and WB. Interestingly, the IHC and WB results revealed that PIN-1 protein levels in lymphoma tissues were drastically increased in GCB DLBCL patients compared to non-GCB DLBCL patients (Figure 1A and B). Furthermore, only 13.2% (5 of 38) of non-GCB DLBCL patient samples were positive for PIN-1, while a much higher proportion of GCB DLBCL patient samples, 51.4% (18 of 35), were positive for PIN-1 (P < 0.001) (Figure 1C). These results indicate that PIN-1 is highly and almost exclusively expressed in GCB DLBCL, allowing us to speculate on the role of PIN-1 in GCB DLBCL.
PIN-1 Depletion Inhibits the Proliferation of GCB DLBCL Cells
Having observed high PIN-1 expression in GCB DLBCL patient samples, we sought to functionally characterize the effects of PIN-1 in GCB DLBCL cells. To this end, we first constructed an shRNA targeting PIN-1 (shPIN-1/2). Our in vitro study focused on investigating the effects of PIN-1 depletion on the proliferation and survival of BJAB cells. According to the results of a Cell Counting Kit-8 (CCK-8) assay, proliferation was markedly lower in PIN-1-knockdown BJAB cells than in shCTRL-transfected BJAB cells (Figure 2A and B). In addition, the same experiment was carried out on HBL-1 cells (ABC DLBCL cells) to detect proliferation with and without PIN-1 depletion. We found that because the basal level of PIN-1 expression in HBL-1 cells was very low (Figure 2C), depletion of PIN-1 did not affect proliferation of these cells (Figure 2D). Furthermore, fluorescence-activated cell sorting (FACS) analysis showed that silencing of PIN-1 alone did not affect the cell cycle in BJAB cells, as shown in Figure 2E. In addition to testing the effect of PIN-1 depletion on the proliferation of GCB DLBCL cell lines, we examined its effect on the survival of primary GCB DLBCL cells. Under in vitro culture conditions, primary GCB DLBCL cells obtained from the tested DLBCL patient samples were cultured for more than 7 days. The proportion of annexin V-positive (apoptotic) cells among these primary DLBCL cells (Figure 2F) increased above basal levels after infection with a PIN-1 shRNA-expressing lentivirus, indicating that the absence of PIN-1 induced apoptosis in primary GCB DLBCL cells. Therefore, we conclude that knockdown of PIN-1 significantly inhibits the proliferation of GCB DLBCL cells.
PIN-1 Defines a GCB DLBCL Subgroup That is Dependent on PI3K/AKT Signaling
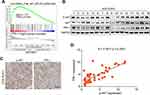
Considering the pivotal role of altered proliferative activity in GCB DLBCL cells, we examined the ERK, NF-κB, PI3K/AKT/mTOR, and E2F pathways, classic signaling pathways associated with tumorigenesis, using gene set enrichment analysis (GSEA) to determine the signaling pathway by which PIN-1 affects GCB DLBCL cell proliferation. Strikingly, PIN-1 was found to be significantly related to the PI3K/AKT/mTOR signaling pathway, as key genes involved in PI3K/AKT/mTOR signaling activation were enriched in patients with elevated levels of PIN-1 expression (Figure 3A). To functionally investigate the role of PIN-1 and determine whether PIN-1 participates in constitutive activation of the PI3K/AKT signaling pathway, we analyzed the levels of p-AKT (a marker of PI3K/AKT activation) and PIN-1 in the context of GCB DLBCL. As expected, the changes in the expression levels of PIN-1 were consistent with those in the expression of p-AKT, as shown by WB of lymphoma tissues from 16 GCB DLBCL lymphoma patients (Figure 3B). Moreover, representative IHC results and the results of statistical analysis revealed that the protein levels of PIN-1 were positively correlated (P<0.0001) with those of p-AKT in lymphoma tumor tissues from 29 GCB DLBCL patients (Figure 3C and D). Taken together, these data indicate that PIN-1 expression is higher in GCB DLBCL patient samples than in non-GCB DLBCL patient samples and is positively associated with p-AKT expression in these samples.
GCB DLBCL is Dependent on PI3K/AKT Signaling
The expression of p-AKT in PIN-1-deficient GCB DLBCL cell lines was examined to determine whether PIN-1 affects PI3K/Akt/mTOR pathway activation in GCB DLBCL cells. The p-AKT protein levels in GCB DLBCL (BJAB) cells were significantly decreased in the absence of PIN-1, as detected by WB (Figure 4A). All-trans retinoic acid (ATRA)-α selectively binds, inhibits and ultimately degrades active PIN-1, thereby exhibiting potent anticancer activity against acute promyelocytic leukemia (APL) and triple-negative breast cancer.13,14 Accordingly, we observed that ATRA treatment decreased the expression of PIN-1 and p-AKT in BJAB cells (Figure 4B).
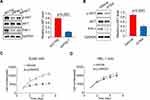
Next, we further explored the clinical prospects of therapeutic targeting of the PI3K/AKT pathway for GCB DLBCL cases in which PIN-1 is highly expressed. To this end, we incubated PIN-1-high GCB DLBCL cells and PIN-1-low ABC DLBCL cells with the pan-PI3K inhibitor LY294002 for 4 days and then determined cell viability. The viability of PIN-1-high BJAB GCB DLBCL cells was significantly reduced by treatment with 5 μM LY294002 (Figure 4C), whereas that of PIN-1-low HBL-1 cells was unaffected (Figure 4D), indicating that LY294002 likely affects cell proliferation through PIN-1. Altogether, our data provide important preclinical evidence supporting an important role of PI3K/AKT signaling in cell survival that can be blocked pharmacologically. Such inhibition might therefore be a therapeutic and preventive approach for GCB DLBCL associated with high PIN-1 expression.
Discussion
It is widely believed that GCB DLBCL patients treated with a standard first-line treatment regimen should subsequently be treated with rituximab plus CHOP (R-CHOP) or with R-CHOP-like chemotherapy, but patients with GCB DLBCL usually have better outcomes than patients with ABC DLBCL following this treatment regimen.
However, even among patients with the GCB subtype of DLBCL, which has a good prognosis, approximately 20% of patients will relapse.15 Importantly, high-grade B cell lymphoma with MYC and BCL-2 and/or BCL-6 rearrangements is associated with very poor outcomes and is usually classified as the GCB subtype of DLBCL by molecular profiling. Multiple studies have shown that PIN-1 plays important roles in the progression and aggressiveness of solid tumors;14 in addition, increasing evidence suggests that it plays the same important roles in leukemia.3,16,17 The experimental results of this study demonstrated that PIN-1 expression was significantly enhanced in GCB DLBCL patient samples and was positively correlated with p-AKT expression. Our study thus used PIN-1 to identify a new germinal center subgroup of DLBCL that is dependent on PI3K/AKT signaling. We investigated the clinical features of PIN-1-positive GCB DLBCL and found that patients with this type of DLBCL showed increased extranodal site infiltration and major lymph node infiltration (15/18, 83%). Our results will be useful in the development of more personalized and targeted management of DLBCL in patients.
The PI3K/Akt/mTOR pathway is highly active in many malignancies, including lymphoid neoplasms. Activation of this pathway in DLBCL may lead to genetic mutations, PTEN deletion, or constitutive activation of upstream regulatory pathways. PTEN is expressed in most ABC subtype samples but is absent in GCB subtype samples, so deletion of the PTEN protein is a marker of the GCB DLBCL subtype.18 In the GCB subtype, PI3K/Akt/mTOR pathway activation or AKT phosphorylation often accompanies the loss of the PTEN protein. In contrast, constitutive phosphorylation of AKT is observed in the ABC DLBCL subtype that is unrelated to PTEN. These results indicate that the mechanism of PI3K/Akt/mTOR pathway activation may differ between DLBCL subtypes and that deletion of the PTEN protein in the GCB subtype may be related to constitutive phosphorylation of AKT.19 In this study, we found that the expression of the prolyl isomerase PIN-1 was elevated in GCB DLBCL patient samples and that high PIN-1 expression was positively correlated with activation of the oncogenic PI3K/AKT pathway in primary patient samples. Depletion of PIN-1 had cytotoxic effects and significantly inhibited proliferation in GCB DLBCL model cell lines by inhibiting PI3K/AKT signaling, indicating a dependence on this pathway in this subgroup of GCB DLBCL. Furthermore, PIN-1 depletion increased apoptosis in primary GCB DLBCL cells. Collectively, our results indicate that PIN-1 defines a PI3K/AKT-dependent GCB DLBCL subgroup that is dependent on PI3K signaling and suggest that PIN-1 is a potential new target for GCB DLBCL therapy.
Functionally, we showed that an established PIN-1/p-AKT relationship may promote GCB DLBCL progression. Indeed, it has recently been demonstrated that PIN-1 downregulation mediated by ATRA, which is used in APL therapy, is correlated with tumor cell growth inhibition in APL model animals and human APL cells in vitro as well as in APL patients.20 Notably, this has also been observed in triple-negative breast cancer, against which ATRA exhibits potent anticancer activity, probably by simultaneously blocking PIN-1 substrate oncogenes and tumor suppressors. For the first time, our findings highlight the isomerase PIN-1 as a potential new therapeutic target for the treatment of GCB DLBCL.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Li S, Young KH, Medeiros LJ. Diffuse large B-cell lymphoma. Pathology. 2018;50(1):74–87. doi:10.1016/j.pathol.2017.09.006
2. Liu Y, Barta SK. Diffuse large B-cell lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am J Hematol. 2019;94(5):604–616. doi:10.1002/ajh.25460
3. Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8(11):904–916. doi:10.1038/nrm2261
4. Liou Y-C, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 2011;36(10):501–514. doi:10.1016/j.tibs.2011.07.001
5. Wulf G, Finn G, Suizu F, Lu KP. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat Cell Biol. 2005;7(5):435–441. doi:10.1038/ncb0505-435
6. Sorrentino G, Comel A, Mantovani F, Del Sal G. Regulation of mitochondrial apoptosis by Pin1 in cancer and neurodegeneration. Mitochondrion. 2014;19:88–96. doi:10.1016/j.mito.2014.08.003
7. Franciosa G, Diluvio G, Gaudio FD, et al. Prolyl-isomerase Pin1 controls Notch3 protein expression and regulates T-ALL progression. Oncogene. 2016;35(36):4741–4751.
8. Cao W-B, Yao J-F, Feng S-Z, et al. BCR-ABL enhances the prolyl isomerase activity of Pin 1 by interacting with DAPK1 in ph(+) ALL. Cancer Med. 2018;7(6):2530–2540. doi:10.1002/cam4.1478
9. Xu -Z-Z, Xia Z-G, Wang A-H, et al. Activation of the PI3K/AKT/mTOR pathway in diffuse large B cell lymphoma: clinical significance and inhibitory effect of rituximab. Ann Hematol. 2013;92(10):1351–1358. doi:10.1007/s00277-013-1770-9
10. Uddin S, Hussain AR, Siraj AK, et al. Role of phosphatidylinositol 3ʹ-kinase/AKT pathway in diffuse large B-cell lymphoma survival. Blood. 2006;108(13):4178–4186. doi:10.1182/blood-2006-04-016907
11. Lu Z, Hunter T. Prolyl isomerase Pin1 in cancer. Cell Res. 2014;24(9):1033–1049. doi:10.1038/cr.2014.109
12. Franciosa G, Diluvio G, Gaudio FD, et al. Prolyl-isomerase Pin1 controls Notch3 protein expression and regulates T-ALL progression. Oncogene. 2016;35(36):4741–4751. doi:10.1038/onc.2016.5
13. Min S-H, Zhou XZ, Lu KP. The role of Pin1 in the development and treatment of cancer. Arch Pharm Res. 2016;39(12):1609–1620. doi:10.1007/s12272-016-0821-x
14. Wei S, Kozono S, Kats L, et al. Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat Med. 2015;21(5):457–466.
15. Friedberg JW. Double-Hit Diffuse Large B-Cell Lymphoma. J Clin Oncol. 2012;30(28):3439–3443. doi:10.1200/JCO.2012.43.5800
16. Pulikkan JA, Dengler V, Peer Zada AA, et al. Elevated PIN1 expression by C/EBP[alpha]-p30 blocks C/EBP[alpha]-induced granulocytic differentiation through c-Jun in AML. Leukemia. 2010;24(5):914–923. doi:10.1038/leu.2010.37
17. Jeong SJ, Ryo A, Yamamoto N. The prolyl isomerase Pin1 stabilizes the human T-cell leukemia virus type 1 (HTLV-1) Tax oncoprotein and promotes malignant transformation. Biochem Biophys Res Commun. 2009;381(2):294–299. doi:10.1016/j.bbrc.2009.02.024
18. Pfeifer M, Lenz G. PI3K/AKT addiction in subsets of diffuse large B-cell lymphoma. Cell Cycle. 2013;12(21):3347–3348. doi:10.4161/cc.26575
19. Pfeifer M, Grau M, Lenze D, et al. PTEN loss defines a PI3K/AKT pathway-dependent germinal center subtype of diffuse large B-cell lymphoma. Procee National Acad Sci. 2013;110(30):12420–12425. doi:10.1073/pnas.1305656110
20. Wei S, Kozono S, Kats L, et al. Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat Med. 2015;21(5):457–466. doi:10.1038/nm.3839
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.