Back to Journals » OncoTargets and Therapy » Volume 14
Targeted-Gene Sequencing and Bioinformatics Analysis of Patients with Pancreatic Mucoepidermoid Carcinoma: A Case Report and Literature Review
Authors Chen Z, Zhang L, Huang J, Ding C, Zhang T , Wan D, Xue L
Received 10 February 2021
Accepted for publication 11 May 2021
Published 3 June 2021 Volume 2021:14 Pages 3567—3581
DOI https://doi.org/10.2147/OTT.S305248
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Zhitao Chen,1,2 Lele Zhang,2,3 Jiacheng Huang,2,3 Chenchen Ding,1 Ting Zhang,1 Dalong Wan,3 Liang Xue3
1Department of Hepatobiliary Surgery, Shulan (Hangzhou) Hospital Affiliated to Zhejiang Shuren University Shulan International Medical College, Hangzhou, People’s Republic of China; 2School of Medicine, Zhejiang University, Hangzhou, People’s Republic of China; 3Department of Hepatobiliary Surgery, First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, People’s Republic of China
Correspondence: Dalong Wan; Liang Xue
Department of Hepatobiliary Surgery, First Affiliated Hospital, School of Medicine, 79# Qingchun Road, Hangzhou City, Zhejiang Province, 310003, People’s Republic of China
Tel +86-571-87236570
Email [email protected]; [email protected]
Background: Primary pancreatic mucoepidermoid carcinoma (MEC) is an extremely rare malignant tumor with unclear etiology and pathogenesis. There are only eleven reported cases in English papers published from 1959 to 2020. MEC generally occurs in the salivary gland, but cases in the pancreas have also been reported. Although being considered as a low-grade indolent carcinoma, pancreatic MEC always invades the surrounding lymph node and metastasizes. The prognosis of pancreatic MEC is unsatisfactory. To date, the genetic alterations, mechanistic relationships among mutated genes and signaling pathways of pancreatic MEC are unclear.
Patient and Methods: This paper presents a case of rare primary pancreatic MEC in a 56-year-old male patient with liver metastasis. Radical surgery of distal pancreatectomy and radiofrequency ablation (RFA) of two liver metastatic lesions is conducted. Targeted-gene sequencing and bioinformatics analysis tools, including STRING, DAVID, cBioPortal, DGidb and Human Protein Atlas database (HPA), are used to clarify the biological functions and features of mutated genes in pancreatic MEC.
Results: Eight gene mutations (TP53, KRAS, ATR, FLI1, FLT4, MAGI2, RBM10, and TNFAIP3) were observed, and a tumor mutation burden (TMB) of 5.6 muts/Mb was calculated in the pancreatic MEC using targeted-gene sequencing. The patient subsequently underwent adjuvant chemotherapy and died three months after surgery. Gene–gene interaction network was constructed, which showed the significant interactions among eight mutated genes. In terms of the functions and pathways of eight gene mutations based on GO and KEGG, 20 tumor-related results are presented in this paper, Furthermore, the biological functions and features of pancreatic MEC are further compared with pancreatic ductal adenocarcinoma.
Conclusion: Pancreatic MEC requires early and effective treatment, and lymph node metastases and multiple organ metastases were unfavorable prognostic factors. Pancreatic MEC can be compared with other pancreatic cancers that have characteristic clinical phenotype, molecular alterations, functional information and enrichment pathway.
Keywords: pancreatic mucoepidermoid carcinoma, targeted-gene sequencing, bioinformatics analysis
Introduction
Mucoepidermoid carcinoma (MEC) is a common primary malignant neoplasm of the salivary gland consisted of intermediate cells, mucinous cells, and epidermoid cells.1 Although MEC can occur at any part of the body, MEC at the pancreas is rare. Related English papers published from 1959 to 2020 are searched using the PubMed database with the search terms “mucoepidermoid carcinoma”, “MEC”, “pancreas” and “pancreatic” and only eleven cases were found. There are no diagnostic criteria of pancreatic MEC and pathologists diagnose based on the morphologic similarity of pancreatic MEC to salivary gland-MEC (Sal-MEC). Some pathologists think that pancreatic MEC is a pathomorphological variant of pancreatic adenosquamous carcinoma.2 However, due to limited reported cases, the clinicopathological features and genetic characteristics of pancreatic MEC remain unclear. Comprehensive gene mutation spectrum and molecular profiling may reveal the reasons for tumorigenesis and clarify the progression of pancreatic MEC.
For pancreatic ductal adenocarcinoma (PDAC), multimodality treatment including radical surgery with adjuvant chemotherapy is preferred and long-term follow-up after the operation is required.3 However, pancreatic MEC is more aggressive than PDAC, neither radical surgical nor chemotherapy contributes to satisfactory prognosis.4 This paper presents a case of a 56-year-old male with pancreatic MEC. In addition to standard histopathological assessments, targeted-gene sequencing is performed. To date, gene mutation spectrum, functional relationship and enrichment pathway of mutated genes in the pancreatic MEC have not been investigated. This paper aims to describe the clinical features, genetic characteristics and clinical management of patients with rare pancreatic MEC. Furthermore, the biological features of pancreatic MEC are further compared with PDAC and expected to provide a better understanding of this disease.
Case Report
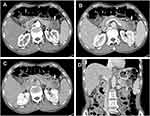
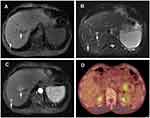
A 56-year-old man with a 1-month history of upper abdominal distension and anorexia was admitted to Shulan (Hangzhou) Hospital on January 9, 2020. Two days ago, the patient went to the local hospital for examination and the abdomen ultrasonic scan showed a hypoechoic focus in the pancreatic tail and isolated mildly hyperechoic foci in the liver. Therefore, the patient was suspected of metastatic tumor. Laboratory tests of the patients showed a modestly elevated leukocyte count of 12.1×109/L (3.5–9.5×109/L); c-reactive protein count of 21.7 mg/dL (≤10 mg/L). The levels of tumor markers are α-fetoprotein (AFP) of 5.0 ng/mL (the normal value: 0.0–20.0 ng/mL), carcinoembryonic antigen (CEA) of 34.8 ng/mL (0.0–5.0 ng/mL), carbohydrate antigen (CA) of 19–9, 2329.8 U/mL (0.0–37.0 U/mL) and CA125, 195.9 U/mL (0.00–35.0 U/mL). Enhanced computed tomography (CT) demonstrated a complex hypoattenuating mass with the size of 4.6×3.7 cm and unclear margins, involving the spleen vessels at the pancreatic tail (Figure 1). Two nodules (one in the S6 of the liver, another in the S8) were suspected to be metastases. Dynamic contrast enhanced magnetic resonance imaging (MRI) of the pancreas was then performed. An irregular heterogeneous mass in the pancreatic tail appeared as hypointense on T1-weighted image and slightly hyperintense on T2-weighted image. Meanwhile, liver lesions appeared as hypointense on T1-weighted image and as hyperintense on T2-weighted image with ring enhanced during the arterial dominant phase of contrast-enhanced MRI, and metastatic lesions could not be excluded (Figure 2A–C). Whole-body 18F-fluorodeoxyglucose (FDG) positron emission tomography (PET)/CT examination revealed a moderate FDG uptake nodule with the size of 4×4.5 cm in front of the pancreatic tail (SUVmax 6.2) and mild FDG uptake nodules in the liver (SUVmax 2.1 and 3.05) with normal scans of the head, neck, chest, and colon (Figure 2D). Subsequently, liver metastases were biopsied with ultrasonography navigation and were interpreted by histopathology as suspected mucinous tumors.
Previous cases of pancreatic MEC prove that chemotherapy is not very effective, and curative surgery with adjuvant chemotherapy is more effective when the mass is resectable.5 Besides, because of frequent upper abdominal distension, the patient preferred curative surgery. Subsequently, the patient underwent distal pancreatectomy accompanied by radiofrequency ablation (RFA) of two liver metastatic lesions. The tumor was gray–yellow and fleshy with a firm texture.
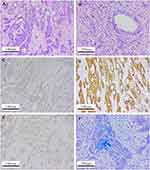
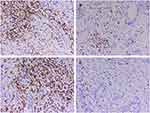
Detailed postoperative pathological examination was performed and it was found that the carcinoma cells consisted of epidermoid cells, intermediate undifferentiated cells and poorly differentiated adenocarcinoma cells with mucous cells in cytoplasm of tumor cell (Figure 3A and B). Immunohistochemical findings indicated that the pancreatic tumor was strong staining with antibodies to P53, CA199, CEA, CK19, CK7, P63, P40 and revealed negative staining for CK20, CD56, CgA, Syn (Figure 3C–F). In addition, the proliferation index by Ki-67 stain was above 80% and demonstrated possibility of a poor prognosis. Based on these findings, the final diagnosis of primary pancreatic MEC and two liver metastases (T3N1M1) was made according to the American Joint Committee on Cancer (AJCC) TNM classification.
Tumor DNA from the pancreatic MEC was tested. The panel covered all exons and selected introns of 448 cancer-related genes. Eight somatic mutations were found, as summarized in Table 1. In addition, the tumor mutation burden (TMB) was calculated as the total number of non-synonymous somatic mutations per megabase (MB) of the genome. The patient had a TMB of 5.6 muts/Mb (Figure 4D), which is higher than PDAC (approximately 1.0 muts/Mb).6,7 Meanwhile, PD-L1 (Programmed cell Death-Ligand 1) immunohistochemistry assays were used to predict PD-1/PD-L1 inhibitor response, which showed PD-L1 expression in pancreatic MEC. Tumor proportion score (TPS) and combined positivity score (CPS) were calculated to evaluate the immunohistochemical expression of PD-L1 based on 22C3 assays and detected in 15% and 8 of the cases, respectively (Figure 4A–C). (This work was accomplished by the OrigiMed biotechnology company)
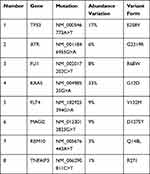 |
Table 1 Shared Gene Alterations in the Pancreatic Mucoepidermoid Carcinoma (MEC) |
The patient developed a postoperative biochemical fistula and recovered after symptomatic treatment and then was discharged on the 28th postoperative day. Considering the positive expression of PD-L1 antibody and genetic characteristics, it was expected that the patient may present a potential sensitivity to immunological therapy. Therefore, pembrolizumab is recommended to the patient as systemic immunotherapy, but the patient refused for economic reasons. Fourteen days after the discharge the patient received a full dose of gemcitabine of 1000 mg/m2 days 1 and 8, and oral oteracil potassium capsules (S-1) 25 mg/m2 twice daily days 1–14 every 3 weeks. Despite the radical surgical treatment and adjuvant chemotherapy, the disease progressed rapidly and examinations revealed multiple abdominal recurrence invading the bile duct. The patient died of liver failure 3 months after the initial operation.
Discussion
MEC is a rare salivary gland-type tumor consisted of epidermoid cells, intermediate undifferentiated cells and poorly differentiated adenocarcinoma cells with mucous cells in cytoplasm of tumor cell. MEC was first reported by Franz in 1959.8 Although the disease generally occurs in the salivary glands, MEC in other organs such as the conjunctiva,9 bronchus,10 thymus,11 and anal canal12 has also been reported. MEC in other organs shares similar histological features with Sal-MEC.13 During the past decade, most MECs are considered to be associated with the translocation involving the CRTC1/3 gene at 19p13 and the MAML2 gene at 11q21 t (11; 19) (q21; p13).14 However, Saeki et al15 have investigated CRTC1/3-MAML2 fusion status in 16 cases of morphologically distinct pancreatic MEC without finding patients with MAML2 fusion and concluded that pancreatic MEC is not a pancreatic counterpart of CRTC1/3-MAML2 fusion gene-related Sal-MEC. Therefore, in this paper, it is speculated that pancreatic MEC has corresponding molecular alterations. However, there are few reported MEC in the digestive system, and pancreatic MEC is extremely rare. English papers published from 1959 to 2020 were searched using the PubMed database with the search terms “mucoepidermoid carcinoma”, “MEC”, “pancreas” and “pancreatic”, and only eleven cases were found.4,5,15–19 Clinicopathologic characteristics of these patients, including age, sex, treatment and prognosis, were collected for a better understanding of pancreatic MEC (Table 2). To date, the gene mutation sites in pancreatic MEC are unknown. The results of the patients’ targeted-gene sequencing tests in this paper may improve clinical management and prognosis of pancreatic MEC.
 |
Table 2 Reported Cases of Pancreatic Mucoepidermoid Carcinoma (MEC) in the English Literature |
Genetic Part
Eight somatic mutations were found, as summarized in Table 1, indicating that pancreatic MEC may be a molecularly heterogeneous disease characterized by eight genetic alterations, namely oncogenic KRAS, FLI1 and FLT4 mutation and inactivation of the tumor suppressors TP53, ATR, MAGI2, RBM10 and TNFAIP3.
According to the WHO classification, pancreatic MEC is regarded as a variant of pancreatic adenosquamous carcinoma with similar morphology. The pancreatic adenosquamous carcinoma is characterized by at least 30% malignant squamous cell carcinoma mixed with ductal adenocarcinoma and has the worst prognosis among different types of pancreatic cancer. TP53 mutations are enriched in squamous subtype tumors. Fang et al20 have further confirmed the TP53 pathway modulates pancreatic adenosquamous carcinoma development. TP53 mutation in the patient in this paper predicts poor prognosis. KRAS mutation plays a vital role in PDAC pathogenesis by encoding a small GTPase that regulates cell proliferation, differentiation, survival, and migration.21 FLI1 and FLT4 have been reported to promote invasion and metastasis of cancer cells, while ATR, MAGI2, RBM10 and TNFAIP3 function as tumor suppressor genes in several kinds of cancers.22–24 These mutations reveal that a few signaling pathways and cellular processes including TP53, ATR, and KRAS, transforming growth factor-β (TGF-β) signaling as well as chromatin remodeling, DNA repair and RNA metabolism, -processing and -decay are closely related to the development of pancreatic MEC.
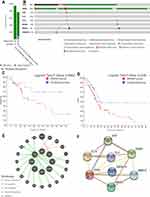
PDAC encompasses more than 85% of pancreatic cancer.25 Gene mutations, including carcinogens and tumor suppressors, are a significant pathological process in the initiation and progression of PDAC. The three major tumor suppressors involved in PDAC are TP53, CDKN2A and SMAD4, and RAS is the most well-known oncogene in PDAC. Subsequently, the mutational status of eight genetic alterations (data from this patient) in PDAC was explored by cBioPortal (www.cbioportal.org) online tool based on TCGA and QCMG databases (mutations data from 562 PDAC samples). The results showed that gene altered in 94.52% of 383 cases (data from QCMG, Nature 2016) and 76.54% of 179 cases (data from TCGA, PanCancer Atlas) (Figure 5A). A hypermutation status of two genes included in pancreatic MEC is observed. In detail, TP53 and KRAS were altered in 81% and 64% of the sequencing data from PAAD samples, respectively (Figure 5B). Unfortunately, none of the database to assess the mutational status of eight genetic alterations in pancreatic adenosquamous carcinoma. In addition, Kaplan–Meier curve results showed that no noticeable discrepancy in disease-free survival of PDAC between the altered group and the unaltered group (Figure 5C, p=0.086). However, genetic alteration was associated with worse overall survival (Figure 5D, p = 0.023) of patients with PDAC.
Furthermore, the eight somatic mutations were analyzed to better understand the biological functions and enrichment pathway. A protein–protein interaction (PPI) information network was constructed with the STRING (https://string-db.org/) to explore the potential interactions among these mutated genes. As expected, a total of 9 nodes and 15 edges were obtained in the PPI network, with a local clustering coefficient of 0.724 (Figure 5E). In addition, GeneMania (http://genemania.org/) results evidenced that the mutated genes in pancreatic MEC were closely connected through genetic and physical interactions (Figure 5F).
To further investigate the functions and mechanisms of the mutated genes, Gene Ontology (GO) functional annotation analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed with the Database for Annotation, Visualization and Integrated Discovery (DAVID) online tools (https://david.ncifcrf.gov/). The p-value <0.01 was the statistically significant criteria, and mutated genes were classified into three functional ontologies, including cellular component (CC), molecular function (MF), and biological processes (BP). As shown in Table 3, the significant results of GO function enrichment analysis presented that mutated genes were mainly clustered in MFs including protein binding, ATP binding, protein self-association, DNA binding, protein phosphatase binding, identical protein binding, protease binding. The BPs functional ontologies presented that the mutated genes were mainly enriched in replicative senescence, cellular response to UV, negative regulation of cell proliferation, Ras protein signal transduction. The category of CCs indicated that mutated genes were primarily clustered in the nucleus, PML body. In addition, the KEGG pathway enrichment analysis presented that mutated genes were mainly enriched in Rap1 signaling pathway, HTLV-I infection, Thyroid cancer, PI3K-Akt signaling pathway, bladder cancer, endometrial cancer, and non-small cell lung cancer. The mutated genes presented different molecular behaviors and peculiar pathway programs. PI3K-Akt signaling pathway was shared by ovarian cancer26 and gastric cancer,27 which are malignancies with a significant clinical response to targeted immunotherapy.
 |
Table 3 Significantly Enriched GO Terms and KEGG Pathways of Mutated Genes |
Genetic mutations serve as targets in precise therapy for cancer. Inhibition of mutated oncogenes (KRAS, FLI1 and FLT4) and reactivate inactivated tumor suppressors (TP53, ATR, MAGI2, RBM10 and TNFAIP3) are two available methods for targeted therapies in patients with pancreatic MEC. To explore potential drugs based on eight mutated genes, the DGidb (https://dgidb.genome.wustl.edu/), a web server, was used to explore the drug–gene interactions of mutated genes. Among the eight mutated genes, five genes, namely KRAS, FLT4, TP53, ATR, and TNFAIP3 showed significant correlations with potential targeted drug (Table S1). According to these data and targeted-gene sequencing, it is concluded that patients with pancreatic MEC present a potential clinical response to Pembrolizumab28,29 (KRAS and TP53 mutation), Nivolumab30 (KRAS mutation), and Atezolizumab30(KRAS mutation).
PD-L1, a ligand expressed by tumor cells, is capable of binding to the PD-1 (programmed death-protein 1). PD-1/PD-L1 signaling pathway plays a vital role in the suppression of antitumor CD8+T-cell function which impairs immunosurveillance of cancer cells.31 So the positive expression of PD-L1 antibody in the patient in this paper not only represents that the immunosuppressive environment contributes to inducing pancreatic MEC tumors efficiently but also shows that the patient may be more expressive and responsive to some immune checkpoint blockers or immune checkpoint blockers combined with other therapeutic strategies.
Clinicopathologic Part
According to the data in Table 2, it can be found that including the case in this paper, eight cases of pancreatic MEC are male (8/12, 67%) and 4 are female (4/12, 33%). The tumor is usually sized between 2.0 and 17.0 cm (mean diameter of 6.8 cm), and the mean age of the patients is 58.6 years (48–69 years). The pancreatic MEC can occur at any site of the pancreas (4 cases in the pancreatic body, 3 cases in the pancreatic head, 3 cases in the pancreatic tail, and 2 cases in the pancreatic body and tail). The tumor cells invade other organs or tissues in a metastasis pattern, often accompanied by lymph node metastasis (5/12 patients) and liver metastasis (6/12 patients). The frequent lymph node metastasis and multiple organ metastases indicate a worse long-term outcome.
Without distinct features, pancreatic MEC is always confused with other types of pancreatic tumors, which is also demonstrated in this paper. A series of preoperative radiological imaging can estimate the location of the mass and the relationship with the surrounding organs and vascular systems. Tumor markers, such as CA-199, CEA, and CA-125, lack specificity and sensitivity. Therefore, the definitive diagnostic criteria of pancreatic MEC depend on histological examination and immunohistochemical staining tests.
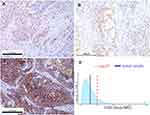
In immunohistochemistry, our patient was positive for P53, CA199, CEA, CK19, CK7, P63, and P40, but negative for CK20, CD56, CgA, and Syn. To gain an improved understanding of MEC in the pancreas by increasing more immunohistochemical study. Abnormal expression of mucins is observed in many adenocarcinoma cells and related to tumor progression, histologic characteristics and prognosis.32,33 We reasoned that pancreatic MEC might show an altered expression of mucins similar to that observed in adenocarcinomas. Subsequently, we investigated pancreatic MEC by immunohistochemistry for MUC1, MUC2, MUC5AC and MUC6 (Figure 6). Meanwhile, the protein expression of MUC1, MUC2, MUC5AC and MUC6 in normal pancreatic tissues and PDAC was also displayed (data from the Human Protein Atlas, https://www.proteinatlas.org/) (Figure 7).
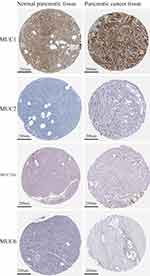 |
Figure 7 The protein expression of MUC1, MUC2, MUC5AC and MUC6 in normal pancreatic tissues and PDAC tissues from the Human Protein Atlas (HPA, https://www.proteinatlas.org/). |
Immunoscore was first proposed by Jérôme Galon in metastatic colon cancer, which studies the densities of CD3+ and cytotoxic CD8+ T cells in the tumor core (TC) and invasive margin (IM) by immunohistochemistry and digital pathology.34,35 In metastatic colon cancer, immunoscore allows more precise definition of patient prognosis than the TNM stage. However, in this patient, the immunohistochemical results demonstrated immune-related T cells enrichment in invasive margin, and tumor core is sparse (intermediate immunoscore, Is2) (Figure 8). We suggested that intermediate immunoscore may be associated with highly aggressive carcinoma and unsatisfactory prognosis. Further accumulation of cases is required to explore a more effective immunoscore.
At present, detailed surgical guidelines regarding effective treatment of pancreatic MEC are unavailable. Curative surgery combined with adjuvant chemotherapy is the main treatment. However, neither radical resection nor chemotherapy has been proved to be effective for pancreatic MEC. Almost all patients with pancreatic MEC died within 6 months and just one patient lived 45 months and the patient in this paper merely lived for 3 months after surgical treatment. The 1-year survival rate in the 12 cases summarized in this paper is 18.1% and the 3-year survival rate is 8.3%. Almost all pancreatic MECs are deteriorated, leading to lymph node metastases and multiple organ metastases, which were poor prognostic factors. More cases are required to explore more effective therapeutic measures of pancreatic MEC.
Conclusion
In conclusion, a case of a patient with rare pancreatic MEC is presented and clinicopathologic and genetic characteristics are discussed based on previous reports. Curative surgery is usually performed in previous cases of pancreatic MEC; however, the prognosis is unsatisfactory. Gene sequencing tests may improve clinical management and prognosis of this disease. More cases are needed to investigate the more effective treatment of pancreatic MEC.
Abbreviations
MEC, mucoepidermoid carcinoma; RFA, radiofrequency ablation; TMB, tumor mutation burden; CEA, carcinoembryonic antigen; PDAC, pancreatic ductal adenocarcinoma; CA, carbohydrate antigen; CT, computed tomography; MRI, magnetic resonance imaging; FDG, fluorodeoxyglucose; PET, positron emission tomography; AJCC, American Joint Committee on Cancer; H&E, hematoxylin and eosin; TPS, Tumor proportion score; CPS, combined positivity score; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Data Sharing Statement
Not applicable.
Ethics Approval and Consent to Participate
Not applicable as this is not a study.
Consent for Publication
Written permission to publish this case report was obtained from the patient.
Acknowledgments
The authors acknowledge the support of the OrigiMed biotechnology company.
Author Contributions
Zhitao Chen: Writing – original draft; Writing – review & editing; Methodology. Lele Zhang: Formal analysis; Methodology; Resources. Jiacheng Huang: Formal analysis; Writing – original draft; Writing. Chenchen Ding: Data curation; Resources. Ting Zhang: Resources. Dalong Wan: Writing – original draft; Writing – review & editing. Liang Xue: Writing – original draft; Formal analysis; Investigation. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
There was no funding for this paper.
Disclosure
The authors declare that they have no competing interests.
References
1. Devaraju R, Gantala R, Aitha H, Gotoor SG. Mucoepidermoid carcinoma. BMJ Case Rep. 2014;2014:
2. Borazanci E, Millis SZ, Korn R, et al. Adenosquamous carcinoma of the pancreas: molecular characterization of 23 patients along with a literature review. World J Gastrointest Oncol. 2015;7(9):132–140. doi:10.4251/wjgo.v7.i9.132
3. Zheng L, Xue J, Jaffee EM, Habtezion A. Role of immune cells and immune-based therapies in pancreatitis and pancreatic ductal adenocarcinoma. Gastroenterology. 2013;144(6):1230–1240. doi:10.1053/j.gastro.2012.12.042
4. Ma R, Yu YQ, Li JT, Peng SY. Mucoepidermoid carcinoma of the pancreas: a case report and a review of literature. J Res Med Sci. 2012;17(9):886–889.
5. Hu HJ, Zhou RX, Liu F, Wang JK, Li FY. You cannot miss it: pancreatic mucoepidermoid carcinoma: a case report and literature review. Medicine (Baltimore). 2018;97(11):e9990. doi:10.1097/MD.0000000000009990
6. Nevala-Plagemann C, Hidalgo M, Garrido-Laguna I. From state-of-the-art treatments to novel therapies for advanced-stage pancreatic cancer. Nat Rev Clin Oncol. 2020;17(2):108–123. doi:10.1038/s41571-019-0281-6
7. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi:10.1038/nature12477
8. Frantz V. Atlas of tumor pathology. section VII-Fascicles 27 and 28. Tumors Pancreas. 1959.
9. Koch KR, Ortmann M, Heindl LM. Conjunctival mucoepidermoid carcinoma. Ophthalmology. 2016;123(3):616. doi:10.1016/j.ophtha.2016.01.016
10. Gieraerts C, Proesmans M, Sauer K, Smet MH, Breysem L. Bronchial mucoepidermoid carcinoma. J Belg Soc Radiol. 2016;100(1):111. doi:10.5334/jbr-btr.1215
11. Roden AC, Erickson-Johnson MR, Yi ES, García JJ. Analysis of MAML2 rearrangement in mucoepidermoid carcinoma of the thymus. Hum Pathol. 2013;44(12):2799–2805. doi:10.1016/j.humpath.2013.07.031
12. Yamamoto M, Hirata K, Tuneyoshi M, et al. Mucoepidermoid carcinoma of the anal canal: a case report and review of the literature. Mol Clin Oncol. 2018;9(5):504–506. doi:10.3892/mco.2018.1706
13. Cipriani NA, Lusardi JJ, McElherne J, et al. Mucoepidermoid carcinoma: a comparison of histologic grading systems and relationship to MAML2 rearrangement and prognosis. Am J Surg Pathol. 2019;43(7):885–897. doi:10.1097/PAS.0000000000001252
14. Pérez-de-oliveira ME, Wagner VP, Araújo ALD, et al. Prognostic value of CRTC1-MAML2 translocation in salivary mucoepidermoid carcinoma: systematic review and meta-analysis. J Oral Pathol Med. 2020;49(5):386–394. doi:10.1111/jop.12970
15. Saeki K, Ohishi Y, Matsuda R, et al. “Pancreatic Mucoepidermoid Carcinoma” Is not a pancreatic counterpart of CRTC1/3-MAML2 fusion gene-related mucoepidermoid carcinoma of the salivary gland, and may more appropriately be termed pancreatic adenosquamous carcinoma with mucoepidermoid carcinoma-like features. Am J Surg Pathol. 2018;42(11):1419–1428. doi:10.1097/PAS.0000000000001135
16. Onoda N, Kang SM, Sugano S, Yamashita Y, Chung YS, Sowa M. Mucoepidermoid carcinoma of the pancreas: report of a case. Surg Today. 1995;25(9):843–847. doi:10.1007/BF00311465
17. Ohtsuki Y, Yoshino T, Takahashi K, Sonobe H, Kohno K, Akagi T. Electron microscopic study of mucoepidermoid carcinoma in the pancreas. Acta Pathol Jpn. 1987;37(7):1175–1182. doi:10.1111/j.1440-1827.1987.tb00435.x
18. Pandey P, Al-Rohil RN, Goldstein JB, et al. Cutaneous metastasis of a mucoepidermoid carcinoma of the pancreas: first reported case. Am J Dermatopathol. 2016;38(11):852–856. doi:10.1097/DAD.0000000000000604
19. Kitagawa S, Suii H, Miyakawa H. Contrast-enhanced endoscopic ultrasonography features of a mucoepidermoid carcinoma of the pancreas. Endosc Ultrasound. 2018;7(5):351–352. doi:10.4103/eus.eus_25_17
20. Fang Y, Su Z, Xie J, et al. Genomic signatures of pancreatic adenosquamous carcinoma (PASC). J Pathol. 2017;243(2):155–159. doi:10.1002/path.4943
21. Luchini C, Paolino G, Mattiolo P, et al. KRAS wild-type pancreatic ductal adenocarcinoma: molecular pathology and therapeutic opportunities. J Exp Clin Cancer Res. 2020;39(1):227. doi:10.1186/s13046-020-01732-6
22. Selvanathan SP, Graham GT, Grego AR, et al. EWS-FLI1 modulated alternative splicing of ARID1A reveals novel oncogenic function through the BAF complex. Nucleic Acids Res. 2019;47(18):9619–9636. doi:10.1093/nar/gkz699
23. Crona DJ, Skol AD, Leppänen VM, et al. Genetic Variants of VEGFA and FLT4 are determinants of survival in renal cell carcinoma patients treated with sorafenib. Cancer Res. 2019;79(1):231–241. doi:10.1158/0008-5472.CAN-18-1089
24. Mazzio EA, Lewis CA, Elhag R, Soliman KF. Effects of Sepantronium Bromide (YM-155) on the Whole Transcriptome of MDA-MB-231 Cells: highlight on Impaired ATR/ATM Fanconi Anemia DNA Damage Response. Cancer Genomics Proteomics. 2018;15(4):249–264. doi:10.21873/cgp.20083
25. Schawkat K, Manning MA, Glickman JN, Mortele KJ. Pancreatic ductal adenocarcinoma and its variants: pearls and perils. Radiographics. 2020;40(5):1219–1239. doi:10.1148/rg.2020190184
26. Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: biological and therapeutic significance. Semin Cancer Biol. 2019;59:147–160. doi:10.1016/j.semcancer.2019.05.012
27. Jia X, Wen Z, Sun Q, et al. Apatinib suppresses the proliferation and apoptosis of gastric cancer Cells via the PI3K/Akt Signaling Pathway. J Buon. 2019;24(5):1985–1991.
28. Dong ZY, Zhong WZ, Zhang XC, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23(12):3012–3024. doi:10.1158/1078-0432.CCR-16-2554
29. Chen N, Fang W, Lin Z, et al. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol Immunother. 2017;66(9):1175–1187. doi:10.1007/s00262-017-2005-z
30. Kim JH, Kim HS, Kim BJ. Prognostic value of KRAS mutation in advanced non-small-cell lung cancer treated with immune checkpoint inhibitors: a meta-analysis and review. Oncotarget. 2017;8(29):48248–48252.
31. Wu Y, Chen W, Xu ZP, Gu W. PD-L1 distribution and perspective for cancer immunotherapy-blockade, knockdown, or inhibition. Front Immunol. 2019;10:2022. doi:10.3389/fimmu.2019.02022
32. Awaya H, Takeshima Y, Yamasaki M, Inai K. Expression of MUC1, MUC2, MUC5AC, and MUC6 in atypical adenomatous hyperplasia, bronchioloalveolar carcinoma, adenocarcinoma with mixed subtypes, and mucinous bronchioloalveolar carcinoma of the lung. Am J Clin Pathol. 2004;121(5):644–653. doi:10.1309/U4WGE9EBFJN6CM8R
33. Chu PG, Weiss LM. Immunohistochemical characterization of signet-ring cell carcinomas of the stomach, breast, and colon. Am J Clin Pathol. 2004;121(6):884–892. doi:10.1309/A09ERYMFR64NERDW
34. Galon J, Pagès F, Marincola FM, et al. The immune score as a new possible approach for the classification of cancer. J Transl Med. 2012;10:1. doi:10.1186/1479-5876-10-1
35. Reichling C, Taieb J, Derangere V, et al. Artificial intelligence-guided tissue analysis combined with immune infiltrate assessment predicts stage III colon cancer outcomes in PETACC08 study. Gut. 2020;69(4):681–690. doi:10.1136/gutjnl-2019-319292
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.