Back to Journals » International Journal of Nanomedicine » Volume 13
Targeted delivery of anti-miR-155 by functionalized mesoporous silica nanoparticles for colorectal cancer therapy
Authors Li Y, Duo Y, Bi J, Zeng X, Mei L, Bao S, He L, Shan A, Zhang Y, Yu X
Received 28 November 2017
Accepted for publication 25 January 2018
Published 1 March 2018 Volume 2018:13 Pages 1241—1256
DOI https://doi.org/10.2147/IJN.S158290
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lei Yang
Yang Li,1,2,* Yanhong Duo,1,3,* Jiangang Bi,1 Xiaowei Zeng,4 Lin Mei,4 Shiyun Bao,1 Lisheng He,5 Aijun Shan,2 Yue Zhang,1 Xiaofang Yu1
1Department of Hepatobiliary and Pancreas Surgery, Second Clinical Medical College of Jinan University, Shenzhen People’s Hospital, Shenzhen 518020, China; 2Department of Emergency, Second Clinical Medical College of Jinan University, Shenzhen People’s Hospital, Shenzhen 518020, China; 3Key Laboratory of Plant Cell Activities and Stress Adaptation, Ministry of Education, School of Life Sciences, Lanzhou University, Lanzhou 730000, China; 4Division of Life and Health Sciences, Graduate School at Shenzhen, Tsinghua University, Shenzhen 518020, China; 5Department of Pathology, Second Clinical Medical College of Jinan University, Shenzhen People’s Hospital, Shenzhen 518020, China
*These authors contributed equally to this work
Introduction: MicroRNA-155 (miR-155) is an oncogenic microRNA, which is upregulated in many human cancers including colorectal cancer (CRC). Overexpression of miR-155 has been found to regulate several cancer-related pathways, and therefore, targeting miR-155 may be an effective strategy for cancer therapy. However, effective and safe delivery of anti-miR-155 to tumors remains challenging for the clinical applications of anti-miR-155-based therapeutics.
Methods: In this study, we explored the expression of miR-155 and the transcription factor nuclear factor kappa B (NF-κB) in CRC tissues and cell lines, and the possible relationship between miR-155 and NF-κB. We further report on anti-miR-155-loaded mesoporous silica nanoparticles (MSNs) modified with polymerized dopamine (PDA) and AS1411 aptamer (MSNs-anti-miR-155@PDA-Apt) for the targeted treatment of CRC.
Results: Results showed that miR-155 is overexpressed in CRC tissues and cell lines, and there is a positive feedback loop between NF-κB and miR-155. Compared to the control groups, MSNs-anti-miR-155@PDA-Apt could efficiently downregulate miR-155 expression in SW480 cells and achieve significantly high targeting efficiency and enhanced therapeutic effects in both in vivo and in vitro experiments. Furthermore, inhibition of miR-155 by MSNs-anti-miR-155@PDA-Apt can enhance the sensitivity of SW480 to 5-fluorouracil chemotherapy.
Conclusion: Thus, our results suggested that MSNs-anti-miR-155@PDA-Apt is a promising nanoformulation for CRC treatment.
Keywords: miR-155, mesoporous silica nanoparticles, AS1411 aptamer, NF-κB, 5-fluorouracil
Introduction
MicroRNAs (miRNAs) are a class of small (18–25 nucleotides) endogenous noncoding RNAs that can regulate gene expressions at the posttranscriptional level.1,2 The aberrant expressions of miRNAs have been proved to be associated with tumor onset, progression, and metastasis.3 Unlike short interfering RNAs (siRNAs) with one specific target mRNA, miRNAs can achieve gene-silencing effect by regulating multiple mRNAs, which make them efficient tools for the treatment of complex multigenic diseases including cancers.4 Restoring miRNA function with miRNA mimics and inhibiting the function of an miRNA with antisense miRNA oligonucleotides (anti-miRs) are the two main strategies to modulate the activity of miRNA.5 Currently, many miRNA-based therapeutics are in the preclinical stage,6 and several miRNA-based drugs have already entered clinical trials for the treatment of cancer and Hepatitis C virus (HCV) infection.7–10 However, effective and safe delivery of anti-miRs or miRNA mimics to the tissue of interest remains a main challenge for the clinical applications of miRNA-based therapeutics.11
MicroRNA-155 (miR-155) is one of the most salient oncogenic microRNA (oncomiR), which is upregulated in many human cancers.12 Overexpression of miR-155 has been found to regulate several cancer-related pathways involved in cell growth, invasion, migration, stemness, and angiogenesis.13 In addition, overexpressed oncogenic miR-155 is also correlated with drug resistance and genome instability.14 Nuclear factor kappa B (NF-κB) is an important transcription factor that regulates the expression of target genes involved in cell growth, apoptosis, angiogenesis, immune and inflammatory response15 and plays an important role in the process of tumor development and progression.16 Previous studies have shown that there is a positive feedback loop between NF-κB and miR-155; therefore, targeting miR-155 may be an effective strategy for the treatment of colorectal cancer (CRC).17,18 Many studies have demonstrated the effectiveness of anti-miR-155 in the treatment of various miR-155-overexpressing tumors, including lymphoma,19,20 liver,21 lung,14 and breast cancers.12
Recently, researchers have explored many viral and nonviral vectors to deliver miRNA mimics or anti-miRs to the target cells. Among these vectors, mesoporous silica nanoparticles (MSNs) have been considered to be promising candidates due to their unique characteristics, including tunable pore structure, large surface area and pore volume, favorable biocompatibility, thermal stability, and easy surface modification.22–25 A series of stimuli-responsive gatekeepers have been developed for the controlled release of drugs from MSNs.26,27 Among them, polymerized dopamine (PDA), formed by the catechol group of dopamine in weak alkaline conditions (pH 8.5), can spontaneously bind to any material surface through covalent and noncovalent interactions.28,29 The PDA film on the surface of MSNs, which serves as a pH-sensitive gatekeeper to controlled release of drug in the acid environment of tumor, can also react with amine- or thiol-terminated active targeting ligands, such as SH-aptamer.30 AS1411 aptamer, the first US Food and Drug Administration-approved aptamer, can specifically bind to nucleolin, which is overexpressed on the cell surface of many tumors including CRC.31–35
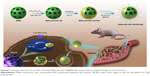
In the present study, we explored the expression of miR-155 and NF-κB in CRC tissues and cell lines and the possible relationship between miR-155 and NF-κB. We further report anti-miR-155-loaded MSNs modified with polydopamine (PDA) and AS1411 aptamer (MSNs-anti-miR-155@PDA-Apt) for the targeted treatment of CRC (as shown in Scheme 1). Furthermore, we evaluate whether the inhibition of miR-155 by MSNs-anti-miR-155@PDA-Apt can enhance the sensitivity of SW480 to 5-FU chemotherapy.
Materials and methods
Materials
Cetyltrimethylammonium bromide (CTAB), tetraethyl orthosilicate (TEOS), 2-(3,4-dihydroxyphenyl) ethylamine (dopamine) hydrochloride, near infrared heptamethine cyanine dyes IR-783, dimethyl sulfoxide (DMSO), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 4′,6-diamidino-2-phenylindole (DAPI), 5′-bromo-2′-deoxyuridine (BrdU) and (3-aminopropyl)-triethoxysilane (APTES) were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). Ammonium fluoride (NH4F) was bought from Aladdin Industrial Co., Ltd. (Shanghai, China). BCA protein assay kit and RIPA lysis buffer were obtained from Beyotime Biotechnology (Jiangsu, China). Dulbecco’s Modified Eagle’s Medium (DMEM), RPMI-1640 cell culture medium, fetal bovine serum (FBS), Trizol Reagent, and Lipofectamine™ 2000 were bought from Thermo Fisher Scientific (Waltham, MA, USA). TaqMan® MicroRNA Reverse Transcription Kit, TaqMan® MicroRNA Assay, and Taqman® Universal PCR Master Mix II were purchased from Thermo Fisher Scientific. 5-Fluorouracil (5-FU) was obtained from Shanghai Xudong Haipu Pharmaceutical Co., Ltd (Shanghai, China). Antibody against BrdU, NF-κB (P65), and β-actin were purchased from Abcam Technology (Cambridge, UK).
Antibody against p-glycoprotein (P-gp) was purchased from Cell Signaling Technology. Horseradish peroxidase (HRP)-linked secondary antibody, rhodamine-conjugated secondary antibody, and fluorescein isothiocyanate-conjugated secondary antibody were acquired from KPL (Gaithersburg, MD, USA). AS1411 aptamer (5′-GGTGGTGGTGGTTGTGGTGGTGGTGG-SH-3′) was synthesized by Sangon Biotech (Shanghai, China). miR-155 (5′UUAAUGCUAAUCGUGAUAGGGGUCCCUAUCACGAUUAGCAUUAAUU3′), anti-miR-155 (5′ACCCCUAUCACGAUUAGCAUUAA3′), NF-κB siRNA (5′GCAGAAGAUGAUCCAUAUUTTAAUAUGGAUCAUCUUCUGCTT3′), siRNA scramble (sense 5′UUCUCCGAACGUGUCACGUTT3′, antisense 5′ACGUGACACGUUCGGAGAATT3′), and FAM (carboxyfluorescein)-anti-miR-155 were synthesized by GenePharma (Jiangsu, China).
Cell lines and cell culture
Human CRC cell lines (SW480, HT-29, SW620, Lovo, and Caco-2) and human normal colon epithelial cell line NCM460 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). SW480, Lovo, Caco-2, SW620, and NCM460 cells were grown in RPMI-1640 medium, and HT-29 cells were grown in DMEM. The cell culture medium was supplemented with 1% streptomycin/penicillin and 10% FBS, and the cells were cultured in a humidified environment containing 5% CO2 at 37°C.
Patients and tissue samples
Paired tumor and nontumor tissue samples of CRC were collected during operation at Shenzhen People’s Hospital between February 2016 and July 2016. The fresh tissue specimens were immediately frozen in liquid nitrogen and subsequently stored at −80°C until the extraction of RNA and protein. The patients’ clinicopathological features (age, gender, tumor differentiation, and TNM classification) were collected from their medical records. All these CRC patients had received no therapy before surgical operation and signed the informed consent. The study was approved by the medical ethics committee of the Second Clinical Medical College of Jinan University.
RNA extraction and quantitative RT-PCR
The tissue was homogenized in liquid nitrogen with a Dounce tissue homogenizer (Biovision, Mountain View, CA, USA). Total RNA was extracted from the tissue and cells using Trizol Reagent according to the manufacturer’s instructions. RNA concentrations were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). Total RNA was reverse transcribed using Taqman® MicroRNA Reverse Transcription Kit and a miRNA-specific looped RT primer. Then the expression of miR-155 was quantified by Taqman® Universal Master Mix II and TaqMan® Assay on the ABI Prism 7300 according to the manufacturer’s protocols.
Western blot analysis
Tissue or cell samples were lysed in RIPA lysis buffer, and the protein concentration was further measured by the BCA assay kit. Equal amounts of proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride (PVDF)membrane. The PVDF membrane was blocked with 5% nonfat dried milk for 2 hours at room temperature and then incubated with rabbit anti-NF-κB (P65) (1:5,000 dilution), mouse anti-P-gp (1:1,000 dilution), or mouse anti-β-actin (1:5,000 dilution) at 4°C overnight. After incubation with HRP-linked secondary antibody (1:3,000 dilution) for 2 hours at room temperature, the protein bands were detected using an enhanced chemiluminescence detection system. Protein levels were normalized to β-actin.
Fabrication of NPs
MSNs were prepared as previously described.36 Briefly, 1.82 g CTAB (5 mmol) and 3 g NH4F (81 mmol) were dissolved in 500 mL of deionized water, and the solution was heated up to 80°C. About 9 mL of TEOS (8.41 g) was then added dropwise into the reaction solution and stirred for another 6 hours. The products were obtained by centrifugation and refluxed in HCl/methanol at 80°C for 24 hours to remove the surfactant template (CTAB). Then, the obtained MSNs were washed with deionized water and dried at 40°C under vacuum.
Preparation of MSNs-NH2
MSNs (200 mg) were refluxed with anhydrous toluene (20 mL) and APTES (0.8 mL, 3.4 mmol) at 120°C for 16 hours. After centrifugation and washing with dichloromethane and diethyl ether, the resulting MSNs–NH2 were dried under vacuum at 60°C for 12 hours.
Preparation of MSNs-anti-miR-155
About 100 mg MSNs–NH2 was added into 5 mL aqueous solution containing four OD anti-miR-155 and stirred for 2 hours at 4°C. The obtained MSNs-anti-miR-155 was then collected by centrifugation and washed with deionized water.
Preparation of MSNs-anti-miR-155@PDA
About 50 mg of MSNs-miR-328 was added into 0.5 mg/mL dopamine hydrochloride in Tris buffer (10 mM, pH 8.5) allowed to react for 3 hours at room temperature under vigorous stirring. Then, the obtained MSNs-anti-miR-155@PDA were collected by centrifugation and washed with deionized water to remove the unpolymerized dopamine.30
Preparation of MSNs-anti-miR-155@PDA-Apt
AS1411 aptamer was covalently conjugated onto the PDA film of MSNs-anti-miR-155@PDA through the -SH group.37
In this study, the above-prepared MSNs-anti-miR-155@PDA was dispersed in Tris-HCl buffer (10 mM, pH 8.5) containing 10 OD AS1411 aptamer and stirred for 3 hours in the dark at room temperature. The resulting material was obtained by centrifugation and washed with deionized water. Finally, the purified MSNs-anti-miR-155@PDA-Apt were resuspended in phosphate-buffered saline (PBS) and stored at 4°C for further use. Fluorescence-labeled MSNs-IR-783@PDA-Apt were prepared according to the same procedure, where IR-783 was used instead of anti-miR-155.
Characterization of the synthesized NPs
The particle size and surface zeta potential of the prepared nanoparticles were measured by a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). The shape and surface morphology of the synthesized NPs were observed by transmission electron microscopy (TEM; FEI Company, Hillsboro, OR, USA). The Fourier transform infrared (FT-IR) spectra of MSNs-anti-miR-155@PDA-Apt was recorded by FT-IR spectrophotometer (Thermo Nicolet, Madison, WI, USA). Thermogravimetric analysis (TGA) of the NPs was performed by Netzsch STA 449 (Netzsch, Selb, Germany).
Finally, the conjugation of AS1411 APt to MSNs@PDA was further confirmed by agarose gel electrophoresis. Samples (MSNs@PDA, MSNs@PDA-Apt, AS1411 aptamer, and DNA ladder) were loaded into a 1% agarose gel. Electrophoresis was carried out at 80 V for 1.5 hours in Tris-Borate EDTA solution. The gel was then stained with ethidium bromide and imaged using a MultiImage™ Light Cabinet (Alpha Innotech Corporation, San Leandro, CA, USA).
In vitro cellular uptake
Fluorescein amidite (FAM)-labeled anti-miR-155 was used to investigate the targeting capability of MSNs-anti-miR-155@PDA-Apt in vitro. SW480 cells were cultured in 12-well plates with an 18 mm cover glasses for 24 hours to allow attachment. Then, the cells were treated with MSNs-FAM-anti-miR-155@PDA and MSNs-FAM-anti-miR-155@PDA-Apt for 6 and 12 hours, respectively. The cells were fixed with 4% paraformaldehyde for 15 minutes, permeabilized with 0.1% Triton X-100–PBS for 10 minutes, and blocked with 3% BSA for 1 hour at room temperature. Subsequently, the cells were incubated with mouse anti-NF-κB (p65) antibody at 4°C overnight and followed by incubation with rhodamine-conjugated secondary antibody for 1 hour at room temperature. After washing with PBS, the cells were counterstained with DAPI for 10 minutes, and fluorescent images were visualized through confocal laser scanning microscopy (CLSM).
The cellular uptake of NPs was also observed by flow cytometry. SW480 cells were seeded in 12-well plates for 24 hours to allow attachment. Then, the cells were treated with MSNs-FAM-anti-miR-155@PDA and MSNs-FAM-anti-miR-155@PDA-Apt for 6 and 12 hours, respectively. Subsequently, the cells were collected and visualized by flow cytometer (BD Biosciences, San Jose, CA, USA).
In vitro cell viability assay
The cytotoxicity of MSNs-anti-miR-155@PDA-Apt against SW480 cells was evaluated by MTT assay, BrdU cell proliferation assay, and colony formation assay.
MTT assay
SW480 or SW480/ADR cells (1×104) were seeded in 96-well plates and cultured overnight to allow attachment. In order to evaluate the cytotoxicity of MSNs-anti-miR-155@PDA-Apt, SW480 cells were treated with MSNs@PDA, MSNs@PDA-Apt, MSNs-anti-miR-155@PDA, and MSNs-anti-miR-155@PDA-Apt for 12, 24, and 48 hours. In order to evaluate the cytotoxicity of combined treatment of MSNs-anti-miR-155@PDA-Apt and 5-FU, SW480 cells were treated with free 5-FU, free 5-FU/MSNs@PDA-Apt, and free 5-FU/MSNs-anti-miR-155@PDA-Apt containing various concentrations of 5-FU ranging from 0 to 100 μM. SW480/ADR cells were treated with free 5-FU, free 5-FU/MSNs@PDA-Apt, and free 5-FU/MSNs-anti-miR-155@PDA-Apt containing various concentrations of 5-FU ranging from 0 to 500 μM. After the above treatments, the medium was replaced with 100 μL of MTT (0.5 mg/mL) at designated time points and further incubated for another 4 hours. Subsequently, the medium was discarded, and 100 μL DMSO was added into each well. The mixture solution was gently vortexed for 10 minutes, and the absorbance was detected at wavelength of 490 nm on a microplate reader.
BrdU cell proliferation assay
SW480 cells (1×104) were plated in 96-well plates with a glass coverslip for 24 hours to allow attachment. Then, the cells were treated with MSNs@PDA, MSNs@PDA-Apt, MSNs-anti-miR-155@PDA, and MSNs-anti-miR-155@PDA-Apt for 6 hours. The medium was replaced with 50 μL 5-fluoiouridine (200 mM) and 1 mL DMEM, and further incubated for another 30 minutes. The cells were fixed with 4% paraformaldehyde for 15 minutes, permeabilized with 0.1% Triton X-100–PBS for 10 minutes, and blocked with 3% BSA for 2 hours at room temperature. Then, the medium was discarded, and the cells were incubated with anti-BrdU mouse monoclonal antibody overnight at 4°C, followed by incubation with fluorescein isothiocynate-conjugated secondary antibody for 1 hour at room temperature. Then, the cells were counterstained with DAPI for 10 minutes, and fluorescent images were acquired through CLSM.
Colony formation assay
SW480 cells were seeded into a six-well plate (1×103 cells/well) and incubated for 24 hours. Then, the cells were treated with MSNs@PDA, MSNs@PDA-Apt, MSNs-anti-miR-155@PDA, and MSNs-anti-miR-155@PDA-Apt in 2 mL culture medium for 72 hours. The medium was replaced with fresh culture medium and further incubated for 7 days. The colonies were stained with 0.1% crystal violet (Sangon Biotech) for 10 minutes and photographed.
SW480 xenograft tumor model
All in vivo studies were carried out in accordance with guidelines for the care and use of laboratory animals, and the protocol was approved by the Administrative Committee on Animal Research in the Second Clinical Medical College of Jinan University. 4–5 weeks old female BALB/c nude mice were purchased from the Medical Experimental Animal Center of Guangdong Province (Guangdong, China). The SW480 tumor xenograft model was established by subcutaneous injection with SW480 cells (2×106) in the flank region. Tumor volume (mm3) = length × (width)2/2.
In vivo imaging and biodistribution
When the tumor volume reached ~130 mm3, the nude mice bearing SW480 tumor xenografts were randomly divided into three groups (n=3 per group). About 100 μL of free IR-783, MSNs-IR-783@PDA, or MSNs-IR-783@PDA-Apt (0.7 mg/kg IR-783 equivalent for NPs) was intravenously injected into nude mice, and the fluorescence signals were captured at 1, 6, 12, and 24 hours postinjection using a whole animal NIR imaging system (Cri Maestro, CRI Inc., Woburn, MA, USA). The excitation wavelength of IR783 was 748 nm, and the emission spectrum of it is 780 nm. At 48 hours after injection, the tumors and major organs (heart, liver, spleen, lung, and kidney) harvested from the sacrificed mice were also imaged by the in vivo imaging system and further quantified by ImageJ (National Institutes of Health, Bethesda, MD, USA).
In vivo antitumor efficacy and systemic toxicity
When the SW480 tumor volume grew up to ~110 mm3, the test mice were randomly divided into seven groups (n=4 per group), and intravenously injected with 100 μL of 1) saline (control), 2) MSNs@PDA-Apt, 3) free anti-miR-155, 4) MSNs-anti-miR-155@PDA, 5) MSNs-anti-miR-155@PDA-Apt, 6) MSNs@PDA-Apt+5-FU, and 7) MSNs-anti-miR-155@PDA-Apt+5-FU at anti-miR-155 dose of 1 nmol and 5-FU dose of 20 mg/kg body weight via the tail vein on 1, 3, and 5 days. Tumor volume and body weight of mice were monitored every 2 days. Fourteen days after the first treatment, the mice were sacrificed, and the harvested tumors were weighed and photographed. Moreover, the slices of tumor and major organs (heart, liver, spleen, lung, and kidney) were further stained with hematoxylin and eosin (H&E) for histological analysis.
Statistical analysis
Data are expressed as mean ± SEM of three independent experiments. SPSS software version 16.0 (SPSS Inc., Chicago, IL, USA) was used to perform the statistical analysis, and P-value <0.05 was considered statistically significant. Differences between groups were analyzed using the Student’s t-test. The correlation between the expression of miR-155 and NF-κB was evaluated by Spearman’s Correlation.
Results and discussion
miR-155 and NF-κB are overexpressed in CRC
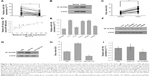
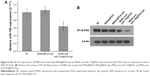
miR-155 has been found to be overexpressed in many malignant tumors and is a promising target for cancer diagnosis, prognosis, and therapy.38 The expression levels of miR-155 were evaluated using qRT-PCR in 30 paired tissue samples of CRC and five CRC cell lines. As shown in Figure 1A and E, miR-155 is significantly overexpressed in CRC tissues compared with adjacent nontumor tissues and in five CRC cell lines (Lovo, HT-29, Caco-2, SW480, and SW620) compared with normal colon epithelial cell line NCM460. Furthermore, we examined the association of miR-155 with the clinicopathologic characteristics in tumor tissues (Table 1). We found that high expression of miR-155 was significantly correlated with poor differentiation and advanced clinical TNM stages, predicting poor prognosis in patients with CRC. MiR-155, functioning as an OncomiR, has multiple targets in human cancers.39 Previous studies show that miR-155 can enhance NF-κB activity by targeting its negative regulators.40,41 Meanwhile, other studies show that miR-155 was found to be upregulated by NF-κB.42,43 Therefore, there may be a positive feedback loop between NF-κB and miR-155.17 In the present study, Western blot analysis showed that NF-κB (P65) proteins were also upregulated in CRC tissues and CRC cell lines (Figure 1B, C, and F), and Spearman’s correlation coefficient test showed that NF-κB (P65) proteins were positively correlated with miR-155 expression in CRC tissues (Figure 1D). In addition, treatment of SW480 cells with miR-155 mimics effectively elevate the protein level of NF-κB (P65) (Figure 1G and H). Downregulation of NF-κB (P65) by siRNA can reduce endogenous miR-155 expression in SW480 cells (Figure 1I). Taken together, these results indicate that there is a positive feedback loop between NF-κB and miR-155, and targeting miR-155 may be an effective strategy for the treatment of CRC. However, specific regulatory mechanisms within this positive feedback loop need further study.
Preparation and characterization of MSNs-anti-miR-155@PDA-Apt
Targeted delivery of anti-miR-155 to tumor cells is a main challenge for the clinical applications of anti-miR-155-based cancer therapy. In this study, we developed anti-miR-155-loaded MSNs modified with PDA and AS1411 aptamer for the targeted treatment of CRC, and the synthetic strategy of MSNs-anti-miR-155@PDA-Apt was schematized in Scheme 1. MSNs were prepared based on our previously reported procedure.36 After loading anti-miR-155 into MSNs through diffusion in an aqueous media, PDA films were coated onto the surfaces of MSNs through the oxidative self-polymerization of dopamine in weak alkaline conditions (pH 8.5). Finally, in order to enhance the targeting effect, SH-terminated AS1411 aptamer (SH-APt) was conjugated on the PDA coating through a Schiff base reaction under weak alkaline conditions.
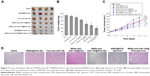
TEM was performed to access the morphology of NPs. As can be observed in Figure 2A, MSNs were uniform and spherical in shape with porous structure. After modification of PDA and AS1411 aptamer, the surface of MSNs-anti-miR-155@PDA-Apt became rough, and a thin film could be obviously visualized on the surface. The size and surface peculiarities of MSNs-anti-miR-155@PDA-Apt measured by DLS are detailed in Table 2. The average diameters of MSNs, MSNs-anti-miR-155, and MSNs-anti-miR-155@PDA-Apt were 125.23±5.46, 130.55±5.65, and 169.43±4.53 nm, respectively, which are all in the appropriate size range for accumulating readily in tumor vasculature due to the enhanced permeability and retention (EPR) effect.44
The zeta potential of MSNs-NH2 decreased from 3.11±4.25 mV to −10.57±3.42 mV after the adsorption of anti-miR-155, which might be due to the negative charge of anti-miR-155. After functionalization with PDA-Apt, the resulting MSNs-anti-miR-155@PDA-Apt had a zeta potential of −15.65±6.26 mV, and the pore structure of MSNs disappeared, indicating that PDA-Apt was successfully conjugated on the surfaces of MSNs.
For quantitative analysis, TGA was performed. As presented in Figure 2B, the weight loss of bare MSNs was only 15% when heated in air atmosphere to 750°C. Nevertheless, after surface functionalization, the weight loss of MSNs@PDA and MSNs@PDA-Apt increased to 21% and 25%, respectively. The amount of PDA film and Apt was calculated to be about 6% and 4%, respectively, indicating the successful modification of PDA film and AS1411 aptamer on the surface of MSNs.
The successful chemical modification was further illustrated using FT-IR spectroscopy (Figure 2C). Peaks at 957 and 1,118 cm−1 were assigned to Si-OH stretch and Si-O-Si symmetric stretch, respectively. Peaks at 1,670 and 3,440 cm−1 could be assigned to physically adsorbed water molecules in MSNs.45,46 C–N stretching vibration of primary amine on MSNs appears at 1,460 cm−1.45 After surface modification with PDA coating and AS1411 aptamer, several new absorption signals appeared. The broad absorbances at 3,395 and 1,740 cm−1 were assigned to the stretching vibrations of N-H/O-H and the overlap of the C=C resonance vibrations, indicating the successful functionalization of PDA coating on MSNs.47,48 Absorption bands at 1,325 cm−1 was mainly assigned to the C/N glycosidic bond of furanoses in DNA oligonucleotides.45 Finally, the conjugation of AS1411 APt to MSNs@PDA was further confirmed by agarose gel electrophoresis. As displayed in Figure 2D, the free AS1411 aptamers showed a band at the molecular weight of 26 bp. However, MSNs did not show any band, and MSNs@PDA-Apt almost stayed at the origin, indicating the successful conjugation of AS1411 aptamers to MSNs@PDA.
In vitro cellular uptake
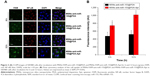
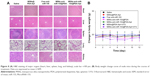
FAM-labeled anti-miR-155 was used to investigate the specific recognition and uptake capacity of MSNs-anti-miR-155@PDA-Apt toward the target cells. SW480 cells were treated with MSNs-FAM-anti-miR-155@PDA and MSNs-FAM-anti-miR-155@PDA-Apt for 6 and 12 hours, and the cellular uptake of nanoparticles was visualized through CLSM. As can be seen from Figure 3A, the green fluorescence of MSNs-FAM-anti-miR-155@PDA-Apt group was much stronger than that of control group (MSNs-FAM-anti-miR-155@PDA) at 6 and 12 hours. This result indicates that MSNs-FAM-anti-miR-155@PDA-Apt has higher cellular uptake, consistent with the results observed by flow cytometry (Figure 3B), which might be attributed to AS1411 Apt-mediated active targeting mechanisms.
In vitro cell viability
The cytotoxicity of MSNs-anti-miR-155@PDA-Apt against SW480 cells was evaluated by MTT assay, BrdU cell proliferation assay, and colony formation assay.
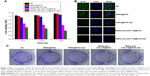
We first evaluated in vitro cytotoxicity of MSNs-anti-miR-155@PDA-Apt using the MTT assay. SW480 cells were treated with MSNs@PDA, MSNs@PDA-Apt, MSNs-anti-miR-155@PDA, and MSNs-anti-miR-155@PDA-Apt for 12, 24, and 48 hours. As shown in Figure 4A, there were no statistically significant differences in viability among blank control group, MSNs@PDA group, and MSNs@PDA-Apt group, indicating that MSNs@PDA-Apt nanoparticles have almost no cytotoxicity on cells. The cytotoxicity of MSNs-anti-miR-155@PDA-Apt was higher than that of MSNs-anti-miR-155@PDA. The BrdU incorporation assays further confirm this result. As shown in Figure 4B, SW480 cells treated with MSNs-anti-miR-155@PDA-Apt have lower BrdU-positive staining than MSNs-anti-miR-155@PDA group.
We then investigated the colony-formation capacity of SW480 cells treated by MSNs-anti-miR-155@PDA. As shown in Figure 4C, there is no obvious difference in the number of colonies among blank control group, MSNs@PDA group, and MSNs@PDA-Apt group. Compared with the other groups, the MSNs-anti-miR-155@PDA-Apt group has the least number of colonies. The above findings show that MSNs@PDA-Apt nanoparticles are biocompatible, and MSNs-anti-miR-155@PDA-Apt have in vitro therapeutic effects on SW480 cells.
Treatment of MSNs-anti-miR-155@PDA-Apt represses the expression of miR-155 and NF-κB (P65) in SW480 cells
We then evaluated whether the MSNs-anti-miR-155@PDA-Apt was able to repress the expression of miR-155 and NF-κB (P65) in SW480 cells. SW480 cells were incubated with MSNs@PDA-Apt and MSNs-anti-miR-155@PDA-Apt for 12 hours, and the expressions of miR-155 and NF-κB (P65) were detected by qRT-PCR and Western blot, respectively. As shown in Figure 5, MSNs-anti-miR-155@PDA-Apt nanoparticles could significantly inhibit the expression of miR-155 and NF-κB (P65) in SW480 cells as compared to blank control as well as MSNs@PDA-Apt, suggesting that MSNs-anti-miR-155@PDA-Apt is highly efficient in delivering anti-miR-155 to SW480 cells.
In vivo imaging and biodistribution
To study in vivo tumor targeting and distribution of MSNs-anti-miR-155@PDA-Apt in nude mice, IR-783, a water-soluble and negatively charged fluorescent dye, was used for near infrared (NIR) imaging.49 About 100 μL of free IR-783, MSNs-IR-783@PDA, and MSNs-IR-783@PDA-Apt (0.7 mg/kg IR-783 equivalent for NPs) were intravenously injected into nude mice bearing SW480 tumor xenografts, and the fluorescence signals were captured by a whole animal NIR imaging system. As shown in Figure 6A and B, the tumor fluorescence intensity of MSNs-IR-783@PDA-Apt group was much stronger than that of free IR-783 and MSNs-IR-783@PDA at 24 hours postinjection. In addition, compared with the other two groups, a significantly enhanced tumor accumulation can also be observed in MSNs-IR-783@PDA-Apt group (Figure 6C and D). The results showed that MSNs-IR-783@PDA-Apt had the best in vivo tumor targeting effects, presumably due to both active targeting of AS1411 aptamer and passive targeting of the EPR effect.
In vivo antitumor effects and systemic toxicity
We further investigated the antitumor effects of MSNs-anti-miR-155@PDA-Apt in nude mice with SW480 tumor xenograft. As shown in Figure 7A–C, compared with saline control group, free anti-miR-155 and MSNs@PDA-Apt treatments had no obvious tumor suppression effect, illustrating that MSNs@PDA-Apt had no antitumor effects and that free anti-miR-155 could be quickly eliminated by nuclease in vivo. However, due to the high CRC selectivity, MSNs-anti-miR-155@PDA-Apt can effectively inhibit the growth of tumor compared with saline control group, which was further confirmed by H&E staining of tumor sections (Figure 7D). The tumor treated with MSNs-anti-miR-155@PDA-Apt exhibited a higher degree of necrosis than the control group.
Body weight and H&E staining of major organs (heart, liver, spleen, lung, and kidney) were used to evaluate the systemic toxicity of MSNs-anti-miR-155@PDA-Apt in vivo. No obvious variation in body weight (Figure 8A) and no noticeable histological changes of the major organs (Figure 8B) were observed in all groups, suggesting that the synthesized NPs have good biocompatibility and no obvious side effects in vivo.
Combined antitumor therapy of MSNs-anti-miR-155@PDA-Apt and 5-FU in vitro and in vivo
Multidrug resistance (MDR) is a major clinical obstacle in the treatment of CRC. Previous studies have shown that overexpression of miR-155 is associated with drug resistance in many human cancers, including CRC.50,55–57 Resistance to 5-FU, one of the commonly used clinical chemotherapeutic agents, occurs in ~90% of CRC patients.51 In the present study, we further evaluated the combined antitumor effect of MSNs-anti-miR-155@PDA-Apt and 5-FU for the treatment of CRC in vitro and in vivo. Cell viability of SW480 and SW480/ADR cells treated with MSNs-anti-miR-155@PDA-Apt and different concentrations of 5-FU were estimated by MTT assay. As shown in Figure 9A and B, SW480/ADR exhibited lower sensitivity to free 5-FU than SW480 cells. Compared with free 5-FU and free 5-FU/MSNs@PDA-Apt group, combined treatment of free 5-FU and MSNs-anti-miR-155@PDA-Apt exhibited higher cytotoxicity on SW480 and SW480/ADR cells, indicating that MSNs-anti-miR-155@PDA-Apt can successfully enhance the sensitivity of SW480 cells to 5-FU. P-gp, an ABC transporter on the MDR tumor cell membrane, has been found to be closely related to chemoresistance in various types of cancer.52–54 As shown in Figure 9C, MSNs-anti-miR-155@PDA-Apt treatment can effectively decrease the expression of P-gp in SW480/ADR cells. Furthermore, we evaluate the combined antitumor therapy in nude mice with SW480 tumor xenograft. As shown in Figures 7 and 9D and E, antitumor therapy of free 5-FU and MSNs-anti-miR-155@PDA-Apt could more effectively inhibit tumor growth than free 5-FU, which was further supported by H&E staining of tumor sections (Figure 7D). The above results indicate that MSNs-anti-miR-155@PDA-Apt can resensitize CRC tumors to 5-FU through downregulation of P-gp.
Conclusion
We found that miR-155 and NF-κB (P65) were significantly increased in CRC tissues, and there is a positive feedback loop between NF-κB and miR-155. We further constructed an anti-miR-155-loaded MSNs modified with PDA and AS1411 aptamer (MSNs-anti-miR-155@PDA-Apt) for the targeted treatment of CRC. The in vitro and in vivo results reveal that MSNs-anti-miR-155@PDA-Apt bioconjugates can efficiently target tumor, leading to efficient gene silencing and tumor growth inhibition, due to both active targeting of AS1411 aptamer and passive targeting of the EPR effect. Moreover, such nanoformulation can resensitize CRC tumors to 5-FU through downregulation of P-gp. Overall, our results demonstrate that the multifunctional nanoplatform reported herein shows great potential as anti-miR15 delivery vehicle for CRC treatment.
Acknowledgment
This work was supported by the Science, Technology & Innovation Commission of Shenzhen Municipality (No JCYJ20160422170206664).
Disclosure
The authors report no conflicts of interest in this work.
References
Ansari AS, Santerre PJ, Uludağ H. Biomaterials for polynucleotide delivery to anchorage-independent cells. J Mater Chem B. 2017;5:7238–7261. | ||
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. | ||
Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: new trends in the development of miRNA therapeutic strategies in oncology. Int J Oncol. 2016;49:5–32. | ||
Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4:e252. | ||
Muthiah M, Park I-K, Cho C-S. Nanoparticle-mediated delivery of therapeutic genes: focus on miRNA therapeutics. Expert Opin Drug Deliv. 2013;10:1259–1273. | ||
Van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 2014;6:851–864. | ||
Beg MS, Brenner AJ, Sachdev J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35:180–188. | ||
Reid G, Kao SC, Pavlakis N, et al. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics. 2016;8:1079–1085. | ||
Querfeld C, Pacheco T, Foss FM, et al. Preliminary results of a phase 1 trial evaluating MRG-106, a synthetic microRNA antagonist (LNA antimiR) of microRNA-155, in patients with CTCL. Blood. 2016;128:1829. | ||
van der Ree MH, de Vree JM, Stelma F, et al. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: a phase 1B, double-blind, randomised controlled trial. Lancet. 2017;389:709–717. | ||
Meng Y, Liu C, Zhao J, et al. An injectable miRNA-activated matrix for effective bone regeneration in vivo. J Mater Chem B. 2016;4:6942–6954. | ||
Jiang S, Zhang H-W, Lu M-H, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. | ||
Ivkovic TC, Voss G, Cornella H, Ceder Y. microRNAs as cancer therapeutics: a step closer to clinical application. Cancer Lett. 2017;407:113–122. | ||
Van Roosbroeck K, Fanini F, Setoyama T, et al. Combining anti-miR-155 with chemotherapy for the treatment of lung cancers. Clin Cancer Res. 2017;23:2891–2904. | ||
Wang S, Liu Z, Wang L, Zhang X. NF-κB signaling pathway, inflammation and colorectal cancer. Cell Mol Immunol. 2009;6:327–334. | ||
Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-[kappa] B as the matchmaker. Nat Immunol. 2011;12:715–723. | ||
Tong L, Yuan Y, Wu S. Therapeutic microRNAs targeting the NF-kappa B signaling circuits of cancers. Adv Drug Deliv Rev. 2015;81:1–15. | ||
Bakirtzi K, Hatziapostolou M, Karagiannides I, et al. Neurotensin receptor 1-microRNA networks modulate colon cancer in humans and mice. Gastroenterology. 2011;141:1749. | ||
Cheng CJ, Bahal R, Babar IA, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. | ||
Babar IA, Cheng CJ, Booth CJ, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109:E1695–E1704. | ||
Zhang M, Zhou X, Wang B, et al. Lactosylated gramicidin-based lipid nanoparticles (Lac-GLN) for targeted delivery of anti-miR-155 to hepatocellular carcinoma. J Control Release. 2013;168:251–261. | ||
Bouchoucha M, Béliveau É, Kleitz F, Calon F, Fortin M-A. Antibody-conjugated mesoporous silica nanoparticles for brain microvessel endothelial cell targeting. J Mater Chem B. 2017;5:7721–7735. | ||
He L, Huang Y, Chang Y, et al. A highly selective dual-therapeutic nanosystem for simultaneous anticancer and antiangiogenesis therapy. J Mater Chem B. 2017;5:8228–8237. | ||
Wang Z, Wu P, He Z, et al. Mesoporous silica nanoparticles with lactose-mediated targeting effect to deliver platinum (IV) prodrug for liver cancer therapy. J Mater Chem B. 2017;5:7591–7597. | ||
Tang F, Li L, Chen D. Mesoporous silica nanoparticles: synthesis, biocompatibility and drug delivery. Adv Mater. 2012;24:1504–1534. | ||
Zhang J, Cai K. Integration of polymers in the pore space of mesoporous nanocarriers for drug delivery. J Mater Chem B. 2017;5:8891–8903. | ||
Song Y, Li Y, Xu Q, Liu Z. Mesoporous silica nanoparticles for stimuli-responsive controlled drug delivery: advances, challenges, and outlook. Int J Nanomedicine. 2017;12:87. | ||
Perikamana SKM, Lee J, Shin YM, et al. Oxygen-dependent generation of a graded polydopamine coating on nanofibrous materials for controlling stem cell functions. J Mater Chem B. 2017;5:8865. | ||
Lee H, Dellatore SM, Miller WM, Messersmith PB. Mussel-inspired surface chemistry for multifunctional coatings. Science. 2007;318:426–430. | ||
Park J, Brust TF, Lee HJ, Lee SC, Watts VJ, Yeo Y. Polydopamine-based simple and versatile surface modification of polymeric nano drug carriers. ACS Nano. 2014;8:3347–3356. | ||
Taghavi S, Nia AH, Abnous K, Ramezani M. Polyethylenimine-functionalized carbon nanotubes tagged with AS1411 aptamer for combination gene and drug delivery into human gastric cancer cells. Int J Pharm. 2017;516:301–312. | ||
Zhu Y-J, Li W-J, Hong Z, Tang A-N, Kong D. Stable, polyvalent aptamer-conjugated near infrared fluorescent nanocomposite for high-performance cancer cell-targeted imaging and therapy. J Mater Chem B. 2017;5:9229–9237. | ||
He L, Zeng L, Mai X, Shi C, Luo L, Chen T. Nucleolin-targeted selenium nanocomposites with enhanced theranostic efficacy to antagonize glioblastoma. J Mater Chem B. 2017;5:3024–3034. | ||
Li L, Hou J, Liu X, et al. Nucleolin-targeting liposomes guided by aptamer AS1411 for the delivery of siRNA for the treatment of malignant melanomas. Biomaterials. 2014;35:3840–3850. | ||
Li X, Yu Y, Ji Q, Qiu L. Targeted delivery of anticancer drugs by aptamer AS1411 mediated Pluronic F127/cyclodextrin-linked polymer composite micelles. Nanomedicine. 2015;11:175–184. | ||
Duo Y, Li Y, Chen C, et al. DOX-loaded pH-sensitive mesoporous silica nanoparticles coated with PDA and PEG induce pro-death autophagy in breast cancer. RSC Adv. 2017;7:39641–39650. | ||
Tao W, Zeng X, Wu J, et al. Polydopamine-based surface modification of novel nanoparticle-aptamer bioconjugates for in vivo breast cancer targeting and enhanced therapeutic effects. Theranostics. 2016;6:470. | ||
Jurkovicova D, Magyerkova M, Kulcsar L, et al. miR-155 as a diagnostic and prognostic marker in hematological and solid malignancies. Neoplasma. 2014;61:241–251. | ||
Higgs G, Slack F. The multiple roles of microRNA-155 in oncogenesis. J Clin Bioinform. 2013;3:17. | ||
Zhou S, Wang Y, Meng Y, et al. In vivo therapeutic success of microRNA-155 antagomir in a mouse model of lupus alveolar hemorrhage. Arthritis Rheumatol. 2016;68:953–964. | ||
Jiang S, Zhang LF, Zhang HW, et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012;31:1985–1998. | ||
Thompson RC, Vardinogiannis I, Gilmore TD. Identification of an NF-κB p50/p65-responsive site in the human MIR155HG promoter. BMC Mol Biol. 2013;14:24. | ||
Gerloff D, Grundler R, Wurm A, et al. NF-κB/STAT5/miR-155 network targets PU. 1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia. 2015;29:535–547. | ||
Yang Y, Wang A, Wei Q, et al. Hyperbranched Polyglycerol-induced porous silica nanoparticles as drug carriers for cancer therapy in vitro and in vivo. ChemistryOpen. 2017;6:158–164. | ||
Zhang B, Luo Z, Liu J, Ding X, Li J, Cai K. Cytochrome c end-capped mesoporous silica nanoparticles as redox-responsive drug delivery vehicles for liver tumor-targeted triplex therapy in vitro and in vivo. J Control Release. 2014;192:192–201. | ||
Wong RC, Chow SY, Zhao S, Fong W-P, Ng DK, Lo P-C. pH-responsive dimeric zinc (II) phthalocyanine in mesoporous silica nanoparticles as an activatable nanophotosensitizing system for photodynamic therapy. ACS Appl Mater Interfaces. 2017;9:23487–23496. | ||
Iqbal Z, Lai EP, Avis TJ. Antimicrobial effect of polydopamine coating on Escherichia coli. J Mater Chem. 2012;22:21608–21612. | ||
Zhang M, Zhang X, He X, Chen L, Zhang Y. A self-assembled polydopamine film on the surface of magnetic nanoparticles for specific capture of protein. Nanoscale. 2012;4:3141–3147. | ||
Song J-T, Yang X-Q, Zhang X-S, Yan D-M, Wang Z-Y, Zhao Y-D. Facile synthesis of gold nanospheres modified by positively charged mesoporous silica, loaded with near-infrared fluorescent dye, for in vivo X-ray computed tomography and fluorescence dual mode imaging. ACS Appl Mater Interfaces. 2015;7:17287–17297. | ||
Pu J, Bai D, Yang X, Lu X, Xu L, Lu J. Adrenaline promotes cell proliferation and increases chemoresistance in colon cancer HT29 cells through induction of miR-155. Biochem Biophys Res Commun. 2012;428:210–215. | ||
Liu B, Liu Y, Zhao L, et al. Upregulation of microRNA-135b and microRNA-182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway. Mol Carcinog. 2017;56:2669–2680. | ||
Fletcher JI, Haber M, Henderson MJ, Norris MD. ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer. 2010;10:147–156. | ||
Ma Y-C, Wang J-X, Tao W, et al. Redox-responsive polyphosphoester-based micellar nanomedicines for overriding chemoresistance in breast cancer cells. ACS Appl Mater Interfaces. 2015;7:26315–26325. | ||
Baguley BC. Multiple drug resistance mechanisms in cancer. Mol Biotechnol. 2010;46:308–316. | ||
Van Roosbroeck K, Fanini F, Setoyama T, et al. Targeting miR-155 to resensitize lung cancer to chemotherapy. European Journal of Cancer. 2016;61:S116. | ||
Meng X, Niu J, Yang Y, et al. Downregulation of miR-155 inhibits proliferation and enhances chemosensitivity to Temozolomide in glioma cells. Int J Clin Exp Pathol. 2017;10(2):1303–1312. | ||
Bayraktar R, Van Roosbroeck K. miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer and Metastasis Reviews. 2018;37:33–44. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.