Back to Journals » Cancer Management and Research » Volume 12
Tanshinone IIA Suppresses Glioma Cell Proliferation, Migration and Invasion Both in vitro and in vivo Partially Through miR-16-5p/Talin-1 (TLN1) Axis
Authors You S, He X, Wang M, Mao L, Zhang L
Received 31 March 2020
Accepted for publication 1 October 2020
Published 6 November 2020 Volume 2020:12 Pages 11309—11320
DOI https://doi.org/10.2147/CMAR.S256347
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Ahmet Emre Eşkazan
Shihao You,1,* Xianghui He,2,* Mei Wang,1 Lina Mao,1 Lu Zhang3
1Department of Neurology, Qingdao Fuwai Cardiovascular Hospital, Qingdao, Shandong, People’s Republic of China; 2Department of Emergency, Qingdao Fuwai Cardiovascular Hospital, Qingdao, Shandong, People’s Republic of China; 3Department of Peripheral Vascular Diseases, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan, Shandong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Lu Zhang Email [email protected]
Background: Tanshinone IIA (TIIA) is one of the active constituents derived from the rhizome of Danshen, a traditional Chinese herbal. Recently, microRNAs (miRNAs) have been suggested to be associated with the anticancer role of TIIA. However, it remains vague of the interaction between miRNAs and TIIA in glioma, a common aggressive brain tumor in humans.
Methods: Expression of miRNA (miR)-16-5p and talin-1 (TLN1) was detected using reverse transcription-quantitative polymerase chain reaction and Western blotting. Cell proliferation, migration and invasion were assessed with cell viability assay, transwell assay, Western blotting, and xenograft tumor experiment. The target binding between miR-16-5p and TLN1 was confirmed by dual-luciferase reporter assay and RNA pull-down assay.
Results: TIIA treatment inhibited cell viability, migration and invasion, and decreased Cyclin D1, matrix metalloproteinase (MMP)-9 and Vimentin expression in glioma T98G and A172 cells both in vitro and in vivo. Thus, TIIA induced anti-glioma role, wherein miR-16-5p was upregulated and TLN1 was downregulated. Moreover, silencing miR-16-5p could abate TIIA-mediated suppression on glioma cell proliferation, migration and invasion in vitro and in vivo. TLN1 overexpression also exerted tumor-promoting effect in TIIA-treated T98G and A172 cells. Mechanically, miR-16-5p could regulate TLN1 expression via target binding, and depleting TLN1 could counteract the inhibitory effect of miR-16-5p knockdown on the curative effect of TIIA in T98G and A172 cells.
Conclusion: TIIA exerted the anti-proliferation, anti-migration and anti-invasion role in glioma cells both in vitro and in vivo partially through regulating miR-16-5p/TLN1 axis.
Keywords: tanshinone IIA, TIIA, miR-16-5p, TLN1, glioma
Introduction
Tanshinone IIA is an important compound existing in the rhizome of Salvia miltiorrhiza Bunge (also called Danshen).1 Naturally, TIIA is a lipophilic diterpene. As a commonly used Chinese herbal medicine, TIIA exerts a good pharmacological activity on nervous system disease, endocrine system disease, and cardiovascular, and cerebrovascular disease, as well as cancer.2,3 Recently, it has been well annotated that TIIA could function a serious of anti-cancer activities in different human cancer cells.4 However, the molecular mechanism of TIIA is not wholly clear yet.
Glioma is one of the most prevalent and aggressive primary tumors in human central nervous system.5 The outcome of glioma patients varies with the tumor stages, and the median survival time of glioblastoma (grade IV) is about one year in spite of receiving effective treatments.6 Chemoradiotherapy combined with surgery has been a standard approach for glioma.7 However, poor curative effects of chemotherapeutic agents always happen due to the blood-brain barrier (BBB) in the brain.8 Luckily, TIIA has been previously announced to be capable to ameliorate BBB permeability and penetrated this barrier.9,10 Therefore, TIIA probably could be a superior anti-glioma drug in clinic.1 Nevertheless, the molecular mechanism of TIIA exerting anti-tumor role remains to be fully disclosed in glioma, especially in glioblastoma.
MicroRNAs (miRNAs) are a class of single-stranded noncoding RNAs with less than 25 nucleotides. Enormous evidences have demonstrated that miRNAs play a critical role in glioma initiation and progression.11,12 It has also been suggested that miRNAs might mediate biosynthesis of tanshinones (including TIIA, tanshinone IIB, tanshinone I, and cryptotanshinone) in the root of Danshen.13 Recently, the regulation of tanshinones on miRNAs has been discovered in different diseases, including cancer.14 Even so, the association between miRNAs and TIIA in many tumors including glioma has been undiscovered yet.
MiRNA (miR)-16-5p, belonging to the miR-15/miR-16 cluster, is a well-known tumor suppressor.15 In glioma, miR-16-5p participates in almost all cell events, such as proliferation, metastasis, apoptosis, radiosensitivity and chemoresistance.16,17 Thus, this miRNA has been suggested as a potential biomarker in the diagnosis and treatment of glioma.18,19 In this study, we intended to investigate the contribution of miR-16-5p to the role of TIIA in malignant behaviors of human glioma cells, as well as its functional downstream target.
Materials and Methods
Cells and Cell Culture
Human glioblastoma cell lines T98G and A172 were purchased from European Collection of Authenticated Cell Cultures (Public health England). Human astrocyte cell line (NHA) was originally from National Infrastructure of Cell Line Resource (Shanghai, China). These cells were cultivated in high-glucose of Dulbecco’s Modified Eagle Medium (DMEM; HyClone, Logan, UT, USA) containing 10% fetal bovine serum. The sterile environment of cell culture was 37°C and 5% CO2.
Cell Transfection
The oligonucleotides miR-16-5p mimic, miR-16-5p inhibitor (anti-miR-16-5p), and siRNA against talin-1 (si-TLN1) were provided by Genepharma (Shanghai, China), as well as their negative controls (miR-NC, anti-miR-NC and si-NC). The overexpression vector pcDNA3.1 (pcDNA) was purchased from Addgene (Cambridge, MA, USA) to construct pcDNA-TLN1 vector. The T98G and A172 cells were seeded in 6-well plate till to 80% confluence prior to cell transfection. And, 50 nM of oligonucleotides or 2 μg of vectors were mixed with Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following to the manufacturer’s protocols. When co-transfection, half nucleotides were utilized. The sequences of si-TLN1 were 5ʹ-UGUUAUUUCCUCCUUUUUCUC-3ʹ (sense) and 5ʹ-GAAAAAGGAGGAAAUAACAGG-3ʹ (antisense), and si-NC was 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ (sense) and 5ʹ-ACGUGACACGUUCGGAGAATT-3ʹ (antisense).
TIIA Treatment
TIIA (T4952) was from Sigma-Aldrich (Louis, MO, USA), and was dissolved in dimethylsulfoxide (DMSO; Sigma-Aldrich) for 3 mg/mL stock solution. The T98G and A172 cells (80% confluence) were subjected with TIIA treatment to a final concentration of 0, 3, 6 and 9 μg/mL. The control cells were treated with 0.3% DMSO.
Cell Viability Assay
Cell viability of T98G and A172 cells was measured by methyl thiazolyl tetrazolium (MTT) assay. The cells and transfected cells (1×104) were re-seeded in 96-well plates overnight, followed with TIIA treatment. A total of 5 wells were utilized for each concentration (3, 6 and 9 μg/mL), and cells in DMEM containing 0.3% DMSO were used as control group (0 μg/mL). The blank wells were also set up by adding DMEM without cells. After 48 h, the adherent cells were incubated with 5 mg/mL of MTT (in DMEM) for 4 h, and then 100 μL of DMSO for 10 min. The absorbance at 490 nm (A490) was measured with a microplate reader. The data were presented as percentage of A490 values normalized to control groups.
Transwell Migration and Invasion Assays
The cell migration and invasion of TIIA-treated T98G and A172 cells were evaluated by transwell chamber (BD Biosciences, Franklin Lakes, NJ, USA). After TIIA treatment for 24 h, transwell assays were performed. For invasion assay, the chamber was pre-coated with matrigel (BD Biosciences). In brief, the matrigel together with DMEM (1:9) was placed on the upper of each chamber in 24-well plates, and this invasion transwell chamber was incubated for 6 h at 37°C. The serum-free cells (5×104) were loaded in the upper vertically and dropwise, while 10% serum-supplemented DMEM was added in the bottom. This transwell invasion system was incubated in 37°C and at 5% CO2 for 48 h. The lower surface of chamber was fixed by methanol, stained with 0.1% crystal violet (in methanol) for 2 h. The invaded cells were photographed in three independent fields (100×) for each well. For migration assay, the chamber was matrigel-free, and the other operations were the same to invasion assay.
Western Blotting
The total cellular proteins from cultured cells and tumor tissues were isolated by radioimmunoprecipitation assay buffer supplemented with protease inhibitor cocktail. Subsequently, 20 μg of protein sample was separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After with 5% non-fat milk blocking, primary antibody incubation, and secondary antibody incubation, membranes carrying protein blots were visualized by Enhanced Chemiluminescent reagents (Millipore). The band density was quantified using image J software and normalized to glyceraldehyde-phosphate dehydrogenase (GAPDH) and indicated control groups. The primary antibodies included anti-Cyclin D1 (#55,506, 1:1000 dilution), anti-MMP-9 (#3852, 1:1000 dilution), anti-Vimentin (#3932, 1:1000 dilution), anti-TLN1 (#4021, 1:1000 dilution) and anti-GAPDH (#97,166, 1:1000 dilution) were provided by Cell Signaling Technology (Beverly, MA, USA).
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
The total cellular RNA in cultured cells and tumor tissues was extracted using an E.Z.N.A. Total RNA Kit (Omega, Norcross, GA, USA). With SuperScript First-strand Synthesis System (Invitrogen) and SYBR Green qPCR Master Mix (Invitrogen), the RNAs were amplified and analyzed on ABI 7500 PCR instrument (Applied Biosystems, Foster City, CA, USA) with special primers. Each sample was tested in triplicate. Every rection was repeated for 4 times. The primers were designed and synthesized by GENEWIZ (Beijing, China). The primers were listed as follows: miR-16-5p, 5ʹ-GCCCCTTCGTCGTTAGA-3ʹ forward and 5ʹ-GTGCAGGGTCCGAGGT-3ʹ reverse; U6, 5ʹ-CTGGTTAGTACTTGGACGGGAGAC-3ʹ forward and 5ʹ-GTGCAGGGTCCGAGGT-3ʹ reverse. The relative miR-16-5p expression was normalized to U6.
Dual-Luciferase Reporter Assay
According to the predicted result, the putative binding nucleotides of miR-16-5p on the 3ʹ untranslated region of TLN1 (TLN1-3ʹUTR) were mutated into the complementary sites using QuikChange Lightning Site-Directed Mutagenesis Kit from Agilent Technologies (Stratagene, La Jolla, CA, USA). Next, the wild type and mutant of TLN1-3ʹUTR (TLN1-3ʹUTR-WT/MUT) were inserted into the luciferase reporter vector pGL4, respectively. After that, T98G and A172 cells were co-transfected with TLN1-3ʹUTR-WT/MUT and miR-16-5p mimic or miR-NC mimic for 48 h. Every transfection group was performed in three wells. The luciferase activities of Renilla and firefly were assessed by Luciferase Assay System Kit System (Promega, Madison, WI, USA).
RNA Pull-Down Assay
MiR-16-5p and miR-NC were labeled with biotin using Biotin RNA Labeling Mix (Roche, Indianapolis, IN, USA), and transcribed with T7/SP6 RN polymerase (Roche); then, biotin-labelled miR-16-5p (bio-miR-16-5p) and bio-miR-NC were treated with RNase-free DNase I (Promega) and RNeasy Mini Kit (Qiagen, Redwood City, CA, USA). After that, M-280 streptavidin magnetic beads were incubated with bio-miR-16-5p or bio-miR-NC for 4 h at room temperature, followed with incubation of cell lysates of T98 and A172 cells. The RNAs bound by bio-miR-16-5p or bio-miR-NC were extracted by E.Z.N.A. Total RNA Kit (Omega) and analyzed by RT-qPCR.
Xenograft Tumor Models
T98G cells were stably transfected with antagomiR-16-5p or antagomiR-NC (Genepharma). Then, T98G cells (5×105) and transfected T98G cells (5×105) were separately subcutaneously injected into the back of NOD-SCID mice (n=4; Vital River Laboratories, Beijing, China). After injection for 7 days, the mice were intraperitoneally injected with TIIA (30 mg/kg) or saline for every two days. During xenograft experiments, the size of tumor was measured every 7 days after cell transplantation, and tumor weight was examined on day 35 after the mice were sacrificed by CO2 asphyxiation. The volume of tumor was calculated using 0.5×Length×Wideth2. The experiment was approved by the Institutional Animal Care and Use Committee of Qingdao Fuwai Cardiovascular Hospital, and carried out following the Institute of Laboratory Animal Resources. Animal studies were performed in compliance with the Guidelines for Care and Use of Laboratory Animals of National Institutes of Health and the Basel Declaration.
Statistical Analysis
Quantitative variables were presented as mean ± standard deviations. The statistical analysis was performed using Student’s t-test (for comparison in two groups) or one-way analysis of variance (ANOVA; for comparison in multiple groups) on GraphPad prism 5 (GraphPad Software, La Jolla, CA, USA). Turkey’s post hoc test was performed after ANOVA. The accepted level of significance was P value <0.05.
Results
TIIA Suppressed Glioma Cell Proliferation, Migration and Invasion in vitro
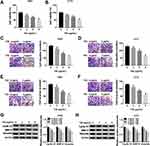
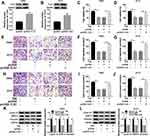
n order to figure out the mechanism of TIIA in glioma progression, T98G and A172 cells were challenged with various doses of TIIA, followed with cell functions analysis. MTT assay illustrated that cell viability of T98G and A172 cells was gradually but significantly attenuated after 3–9 μg/mL of TIIA treatment for 48 h (Figure 1A and B); meanwhile, expression of Cyclin D1 (a key biomarker of cell cycle progression) was decreased in TIIA-disposed T98G and A172 cells (Figure 1G and H). Transwell assays illuminated that migrated cells (Figure 1C and D) and invaded cells (Figure 1E and F) were diminished with the treatment of TIIA ranging from 3 μg/mL to 9 μg/mL for 24 h. Besides, cell metastasis-related proteins MMP-9 and Vimentin were lowly expressed in TIIA-disposed T98G and A172 cells, as well (Figure 1G and H). These outcomes demonstrated TIIA could suppress glioma cell proliferation, migration and invasion in vitro, suggesting a therapeutic effect of TIIA in glioma.
TIIA Induced miR-16-5p Upregulation and TLN1 Downregulation in Glioma Cells
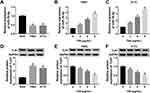
Next, we sought to detect the dysregulation of informative genes induced by TIIA in glioma. According to RT-qPCR and Western blotting analysis, expression of miR-16-5p was lower, and TLN1 was higher in glioma T98G and A172 cells comparing to that in NHA cells (Figure 2A and D). In response to TIIA stimulation, miR-16-5p was distinctively upregulated in T98G and A172 cells in a certain of dose-dependent manner (Figure 2B and C); in the same time, TLN1 was apparently inhibited in TIIA-disposed T98G and A172 cells (Figure 2E and F). These data indicated that miR-16-5p and TLN1 might participate in the pharmacology of TIIA in glioma.
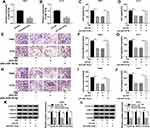
Silencing of miR-16-5p Improved Cell Proliferation, Migration and Invasion in Glioma Cells in vitro Under TIIA Treatment
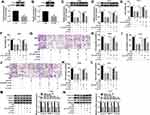
To further validate the role of miR-16-5p in TIIA-treated glioma cells, oligomers anti-miR-16-5p and anti-NC were transfected into T98G and A172 cells, followed with TIIA treatment. As shown, miR-16-5p expression was markedly silenced in anti-miR-16-5p-transfected cells (Figure 3A and B). Due to miR-16-5p silencing, a recovery of cell proliferation was noticed in TIIA-disposed T98G and A172 cells, as evidenced by increased cell viability (Figure 3C and D) and Cyclin D1 expression (Figure 3K and L). Moreover, the depressed cell migration and invasion of TIIA-disposed T98G and A172 cells were improved in the presence of anti-miR-16-5p, as depicted by the increase of transwell migrated and invaded cells (Figure 3E–J), as well as elevated MMP-9 and Vimentin expression (Figure 3K and L). These results showed downregulation of miR-16-5p could hinder the tumor-suppressive role of TIIA in glioma in vitro.
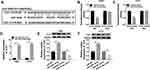
Overexpression of TLN1 Rescued Cell Proliferation, Migration and Invasion in Glioma Cells in vitro Under TIIA Treatment
Similarly, the influence of TLN1 on TIIA-mediated role in malignant behaviors of glioma cells was further confirmed. T98G and A172 cells were transfected with pcDNA3.1-TLN1 vectors or the empty pcDNA vectors prior to 9 μg/mL of TIIA treatment. After then, we identified a high transfection efficiency of TLN1 in TIIA-disposed cells (Figure 4A and B). Cell viability was inhibited by TIIA treatment at 48 h, and this effect was restored by pre-transfection of TLN1 (Figure 4C and D). Cell migration and invasion were suppressed in TIIA-disposed T98G and A172 cells at 24 h, which was improved when TLN1 was pre-overexpressed (Figure 4E–J). TIIA-induced inhibition on Cyclin D1, MMP-9 and Vimentin expression was rescued due to pcDNA3.1-TLN1 transfection (Figure 4K and L). These results also showed that knockdown of TLN1 could impair TIIA role in glioma cells in vitro by inhibiting proliferation, migration and invasion.
There Was a Direct Binding Relationship Between miR-16-5p and TLN1 in Glioma Cells
Herewith, we further identified the interaction between miR-16-5p and cytoskeleton protein TLN1. TLN1 had been known as a molecule involved in cell-cell adhesion and cell motility.20 Bioinformatics algorithms of starbase (http://starbase/hsa_miR-16-5p-TLN1) predicted a potential binding sequence of miR-16-5p on TLN1 3ʹUTR, as presented in Figure 5A. Dual-luciferase reporter assay was conducted to confirm this hypothesis. As a result, the normalized luciferase activity of vectors containing TLN1-3ʹUTR-WT was attenuated in T98G and A172 cells introduced with miR-16-5p mimic (Figure 5B and C). Moreover, RNA pull-down assay indicated an enrichment of TLN1 by bio-miR-16-5p in both T98G and A172 cells (Figure 5D). Western blotting assay monitored TLN1 expression, and manifested that TLN1 protein level was higher in anti-miR-16-5p-transfected T98G and A172 cells, and was lower in miR-16-5p mimic-transfected T98G and A172 cells (Figure 5E and F). These data suggested a direct binding relationship between these two genes in glioma cells.
TLN1 Downregulation Abrogated the Tumor-Promoting Role of miR-16-5p Silencing in TIIA-Disposed Glioma Cells in vitro
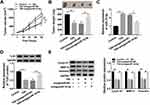
To test whether TIIA exerted anti-glioma role via miR-16-5p/TLN1 axis, we carried out a series of rescue experiments in glioma cells. T98G and A172 cells were treated with 9 μg/mL of TIIA following the transfection of anti-miR-16-5p alone or together with si-TLN1 or si-NC. RT-qPCR identified the high knockdown efficiency of si-TLN1 in both T98G and A172 cells (Figure 6A and B). TLN1 expression was highly induced when miR-16-5p was forcedly silenced via transfection, and this upregulation was further neutralized in the presence of si-TLN1 (Figure 6C and D). Expectedly, the recovery functions of miR-16-5p downregulation on cell viability (Figure 6E and F), migration and invasion (Figure 6G–L) and expression of Cyclin D1, MMP-9 and Vimentin (Figure 6M and N) in TIIA-disposed cells were relieved by si-TLN1 transfection. Thus, we concluded that there might be a miR-16-5p/TLN1 regulatory axis underlying TIIA suppressing glioma in vitro.
Silencing miR-16-5p Attenuated the Suppression of TIIA on Glioma Tumor Growth, Migration and Invasion in vivo via Upregulating TLN1
The xenograft tumor experiment was further launched to explore the TIIA/miR-16-5p/TLN1 route in gliomagenesis in vivo. As shown in Figure 7A and B, T98G cells exhibited a tumorigenicity in NOD-SCID mice, and TIIA treatment dramatically decreased the size and weight of xenograft tumors, whereas pre-transfection of antagomiR-16-5p promoted the growth of TIIA-treated neoplasm (Figure 7A and B). Meanwhile, TIIA upregulated miR-16-5p expression, and downregulated TLN1 expression in T98G-induced neoplasm tissues, which were partially reversed by miR-16-5p stable knockdown (Figure 7C and D). Furthermore, Cyclin D1, MMP-9 and Vimentin expression levels were also suppressed by TIIA treatment in xenograft tumors, and this suppression was attenuated with antagomiR-16-5p transfection (Figure 7E). These findings demonstrated an anti-glioma role of TIIA in glioma cells in vivo by modulating miR-16-5p and TLN1.
Discussion
TIIA was one of the effective compounds derived from the root of Danshen. Di Cesare Mannelli et al21 noticed that Danshen, TIIA and cryptotanshinone could separately attenuate chemotherapy-induced neuropathic pain in animal model, and TIIA could also reveal anti-proliferation/cytotoxicity activity in glioblastoma LN-229 and U-87 MG cells. It had been reported that TIIA exerted its anticancer activity in glioma through modulating growth, differentiation, migration, apoptosis, and autophagy via signaling pathways.22–24 Moreover, Yang et al25 discovered that its anticancer activity in glioma stem cells as well, which was attributed to inhibiting cell growth and stemness, and enhancing apoptosis and differentiation both in vitro and in vivo. Whereas the field of miRNAs regulation in TIIA role was barren in glioma. In this study, we attempted to figure out the role of miR-16-5p and TLN1 in cell proliferation, migration and invasion of TIIA-treated glioblastoma T98 and A172 cells both in vitro and in vivo.
There were quite a few of researches demonstrating miR-16-5p as tumor suppressor in glioma. Consistent with previous studies,16,17 we observed a downregulation of miR-16-5p in human glioblastoma cells. While miR-16-5p level began to be upregulated in response to TIIA treatment, which suggested that TIIA exerting anti-glioma role was partially attributed to miR-16-5p upregulation. In addition, several other miRNAs had been recommended to participate in the anticancer role of TIIA, such as miR-155, miR-122 and miR-30b.26–28 Here, we showed that knockdown of miR-16-5p could counterbalance the suppression of TIIA on cell viability, migration, invasion, and tumor growth, as well as expression of biomarkers of cell proliferation, migration and invasion (Cyclin D1, MMP-9 and Vimentin). The effect of miR-16-5p on Cyclin D1, MMP-9 and Vimentin expression was in accordance with other findings.17,29,30 Besides, miR-16-5p had been implicated with radioresistance and chemoresistance in glioma. For example, Chaudhry et al31 examined the miRNAs expression pattern in carcinogenesis of glioblastoma cells under ionizing radiation therapy and found that miR-16-5p was deregulated in irradiated M059J and M059K cells. Han et al32 validated that miR-16 level was even lower in temozolomide-resistant U251MG cells (U251MG/TR) than the parent cells, and overexpressing miR-16-5p via transfection could sensitize U251MG/TR cells to temozolomide.
Multiple downstream functional genes were directly modulated by miR-16-5p, including Cyclin D1 and Cyclin E1,29 Bmi1,33 Bcl2 and the NF-κB1.17 Herewith, we confirmed TLN1 to be a novel target gene of miR-16-5p in glioma. By the way, TLN1 was correlated with a metastatic phenotype of malignant tumors, such as breast cancer, hepatocellular carcinoma, and prostate cancer.34–36 In glioma, TLN1 had been declared to be highly expressed in bevacizumab-resistant orthotopic xenograft glioma tumor.37 However, TLN1 expression in glioma cells remained unclear comparing to normal brain cells. Here, our data indicated that TLN1 was highly expressed in human glioblastoma T98 and A172 cells, and this upregulation was decreased due to TIIA treatment. Previous findings had also clarified the role of TLN1 in maintaining tensional homeostasis and reversing bevacizumab resistance.20,37 In cellular functions, loss of TLN1 could diminish acquired malignant progressions of glioma cells, such as clonogenic growth, invasion, migration, epithelial–mesenchymal transition, and cancer stem cell properties.37 TLN1-deficient glioma cells were impaired in the spreading and motility. 20 In this study, overexpression of TLN1 could promote cell viability, migration, invasion and expression of Cyclin D1, MMP-9 and Vimentinin glioma cells in spite of TIIA treatment in vitro; on the contrary, TLN1 silencing attenuated above cell behaviors of TIIA-treated T98 and A172 cells with miR-16-5p downregulation. Thus, we might conclude a TIIA/miR-16-5p/TLN1 pathway underlying anti-proliferation, anti-migration and anti-invasion role of TIIA in glioma. By the way, the signaling pathways such as AP-1, integrin, and ERK1/2 signals35,38,39 behind this axis remained unknown.
Taken together, we explored that miR-16-5p upregulation and TLN1 downregulation were accompanied with TIIA treatment in T98 and A172 cells. Downregulation of miR-16-5p and overexpression of TLN1 could separately counterbalance the suppressive role of TIIA in cell proliferation, migration and invasion in glioblastoma cells in vitro and in vivo. TLN1 was identified as one novel target gene of miR-16-5p via directly binding. These data suggested a potential miR-16-5p/TLN1 axis underlying the anti-glioma role of TIIA.
Funding
This work was supported by Exploration and Advantage of Stroke Unit Construction in Traditional Chinese Medicine (GS-FWYY-2019-004).
Disclosure
The authors declare that they have no financial or non-financial conflicts of interest for this work.
References
1. Javed S, Tariq A, Ahmed T, et al. Tanshinones and mental diseases: from chemistry to medicine. Rev Neurosci. 2016;27(8):777–791. doi:10.1515/revneuro-2016-0012
2. Xu S, Liu P. Tanshinone II-A: new perspectives for old remedies. Expert Opin Ther Pat. 2013;23(2):149–153. doi:10.1517/13543776.2013.743995
3. Tian XH, Wu JH. Tanshinone derivatives: a patent review (January 2006–September 2012). Expert Opin Ther Pat. 2013;23(1):19–29. doi:10.1517/13543776.2013.736494
4. Zhang Y, Jiang P, Ye M, Kim SH, Jiang C, Lu J. Tanshinones: sources, pharmacokinetics and anti-cancer activities. Int J Mol Sci. 2012;13(10):13621–13666. doi:10.3390/ijms131013621
5. Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392(10145):432–446. doi:10.1016/S0140-6736(18)30990-5
6. Darefsky AS, King JT
7. Weller M, van den Bent M, Tonn JC, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18(6):e315–e329. doi:10.1016/S1470-2045(17)30194-8
8. Chen MH, Jenh YJ, Wu SK, Chen YS, Hanagata N, Lin FH. Non-invasive photodynamic therapy in brain cancer by use of Tb(3+)-Doped LaF3 nanoparticles in combination with photosensitizer through X-ray irradiation: a Proof-of-Concept Study. Nanoscale Res Lett. 2017;12(1):62. doi:10.1186/s11671-017-1840-3
9. Tang C, Xue H, Bai C, Fu R, Wu A. The effects of tanshinone IIA on blood-brain barrier and brain edema after transient middle cerebral artery occlusion in rats. Phytomedicine. 2010;17(14):1145–1149. doi:10.1016/j.phymed.2010.03.017
10. Lam BY, Lo AC, Sun X, Luo HW, Chung SK, Sucher NJ. Neuroprotective effects of tanshinones in transient focal cerebral ischemia in mice. Phytomedicine. 2003;10(4):286–291. doi:10.1078/094471103322004776
11. Ahir BK, Ozer H, Engelhard HH, Lakka SS. MicroRNAs in glioblastoma pathogenesis and therapy: a comprehensive review. Crit Rev Oncol Hematol. 2017;120:22–33. doi:10.1016/j.critrevonc.2017.10.003
12. Guo Y, Hong W, Wang X, et al. MicroRNAs in microglia: how do MicroRNAs affect activation, inflammation, polarization of microglia and mediate the interaction between microglia and glioma? Front Mol Neurosci. 2019;12:125. doi:10.3389/fnmol.2019.00125
13. Xu X, Jiang Q, Ma X, et al. Deep sequencing identifies tissue-specific microRNAs and their target genes involving in the biosynthesis of tanshinones in Salvia miltiorrhiza. PLoS One. 2014;9(11):e111679. doi:10.1371/journal.pone.0111679
14. Lin X, Qureshi MZ, Romero MA, et al. Regulation of signaling pathways by tanshinones in different cancers. Cell Mol Biol (Noisy-Le-Grand). 2017;63(9):53–58. doi:10.14715/cmb/2017.63.9.10
15. Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17(2):215–220. doi:10.1038/cdd.2009.69
16. Krell A, Wolter M, Stojcheva N, et al. MiR-16-5p is frequently down-regulated in astrocytic gliomas and modulates glioma cell proliferation, apoptosis and response to cytotoxic therapy. Neuropathol Appl Neurobiol. 2019;45(5):441–458. doi:10.1111/nan.12532
17. Yang TQ, Lu XJ, Wu TF, et al. MicroRNA-16 inhibits glioma cell growth and invasion through suppression of BCL2 and the nuclear factor-kappaB1/MMP9 signaling pathway. Cancer Sci. 2014;105(3):265–271. doi:10.1111/cas.12351
18. Ye X, Wei W, Zhang Z, et al. Identification of microRNAs associated with glioma diagnosis and prognosis. Oncotarget. 2017;8(16):26394–26403. doi:10.18632/oncotarget.14445
19. Ivo D’Urso P, Fernando D’Urso O, Damiano Gianfreda C, Mezzolla V, Storelli C, Marsigliante S. miR-15b and miR-21 as circulating biomarkers for diagnosis of glioma. Curr Genomics. 2015;16(5):304–311. doi:10.2174/1389202916666150707155610
20. Sen S, Ng WP, Kumar S. Contributions of talin-1 to glioma cell-matrix tensional homeostasis. J R Soc Interface. 2012;9(71):1311–1317. doi:10.1098/rsif.2011.0567
21. Di Cesare Mannelli L, Piccolo M, Maione F, et al. Tanshinones from salvia miltiorrhiza bunge revert chemotherapy-induced neuropathic pain and reduce glioblastoma cells malignancy. Biomed Pharmacother. 2018;105:1042–1049. doi:10.1016/j.biopha.2018.06.047
22. Wang J, Wang X, Jiang S, et al. Growth inhibition and induction of apoptosis and differentiation of tanshinone IIA in human glioma cells. J Neurooncol. 2007;82(1):11–21. doi:10.1007/s11060-006-9242-x
23. Dong W, Zhang Y, Chen X, Jia Y. High-dose tanshinone IIA suppresses migration and proliferation while promoting apoptosis of astrocytoma cells via notch-1 pathway. Neurochem Res. 2018;43(9):1855–1861. doi:10.1007/s11064-018-2601-0
24. Ding L, Ding L, Wang S, et al. Tanshinone IIA affects autophagy and apoptosis of glioma cells by inhibiting phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin signaling pathway. Pharmacology. 2017;99(3–4):188–195. doi:10.1159/000452340
25. Yang L, Guo H, Dong L, Wang L, Liu C, Wang X. Tanshinone IIA inhibits the growth, attenuates the stemness and induces the apoptosis of human glioma stem cells. Oncol Rep. 2014;32(3):1303–1311. doi:10.3892/or.2014.3293
26. Tu J, Xing Y, Guo Y, Tang F, Guo L, Xi T. TanshinoneIIA ameliorates inflammatory microenvironment of colon cancer cells via repression of microRNA-155. Int Immunopharmacol. 2012;14(4):353–361. doi:10.1016/j.intimp.2012.08.015
27. Zhang H-S, Zhang F-J, Li H, Liu Y, Du G-Y, Huang Y-H. Tanshinone IIA inhibits human esophageal cancer cell growth through miR-122-mediated PKM2 down-regulation. Arch Biochem Biophys. 2016;598:50–56. doi:10.1016/j.abb.2016.03.031
28. Ren X, Wang C, Xie B, et al. Tanshinone IIA induced cell death via miR30b-p53-PTPN11/SHP2 signaling pathway in human hepatocellular carcinoma cells. Eur J Pharmacol. 2017;796:233–241. doi:10.1016/j.ejphar.2016.11.046
29. Hong L, Qing O, Ji Z, et al. Downregulation of miR-16 via URGCP pathway contributes to glioma growth. Sci Rep. 2017;7(1):13470. doi:10.1038/s41598-017-14035-2
30. Wang Q, Li X, Zhu Y, Yang P. MicroRNA-16 suppresses epithelial-mesenchymal transition-related gene expression in human glioma. Mol Med Rep. 2014;10(6):3310–3314. doi:10.3892/mmr.2014.2583
31. Chaudhry MA, Sachdeva H, Omaruddin RA. Radiation-induced micro-RNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol. 2010;29(9):553–561. doi:10.1089/dna.2009.0978
32. Han J, Chen Q. MiR-16 modulate temozolomide resistance by regulating BCL-2 in human glioma cells. Int J Clin Exp Pathol. 2015;8(10):12698–12707.
33. Chen X, Li D, Gao Y, et al. Long intergenic noncoding RNA 00152 promotes glioma cell proliferation and invasion by interacting with MiR-16. Cell Physiol Biochem. 2018;46(3):1055–1064. doi:10.1159/000488836
34. Singel SM, Cornelius C, Batten K, et al. A targeted RNAi screen of the breast cancer genome identifies KIF14 and TLN1 as genes that modulate docetaxel chemosensitivity in triple-negative breast cancer. Clin Cancer Res. 2013;19(8):2061–2070. doi:10.1158/1078-0432.CCR-13-0082
35. Chen P, Lei L, Wang J, et al. Downregulation of Talin1 promotes hepatocellular carcinoma progression through activation of the ERK1/2 pathway. Cancer Sci. 2017;108(6):1157–1168. doi:10.1111/cas.13247
36. Jin JK, Tien PC, Cheng CJ, et al. Talin1 phosphorylation activates beta1 integrins: a novel mechanism to promote prostate cancer bone metastasis. Oncogene. 2015;34(14):1811–1821. doi:10.1038/onc.2014.116
37. Kang W, Kim SH, Cho HJ, et al. Talin1 targeting potentiates anti-angiogenic therapy by attenuating invasion and stem-like features of glioblastoma multiforme. Oncotarget. 2015;6(29):27239–27251. doi:10.18632/oncotarget.4835
38. Lu DY, Leung YM, Cheung CW, Chen YR, Wong KL. Glial cell line-derived neurotrophic factor induces cell migration and matrix metalloproteinase-13 expression in glioma cells. Biochem Pharmacol. 2010;80(8):1201–1209. doi:10.1016/j.bcp.2010.06.046
39. Carbonell WS, DeLay M, Jahangiri A, Park CC, Aghi MK. beta1 integrin targeting potentiates antiangiogenic therapy and inhibits the growth of bevacizumab-resistant glioblastoma. Cancer Res. 2013;73(10):3145–3154. doi:10.1158/0008-5472.CAN-13-0011
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.