Back to Journals » International Journal of General Medicine » Volume 14
Systemic Profiling of KDM5 Subfamily Signature in Non-Small-Cell Lung Cancer
Authors Hao F
Received 3 August 2021
Accepted for publication 14 October 2021
Published 27 October 2021 Volume 2021:14 Pages 7259—7275
DOI https://doi.org/10.2147/IJGM.S329733
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Fang Hao
Department of Oncology, Tianjin Medical University General Hospital, Tianjin, People’s Republic of China
Correspondence: Fang Hao Email [email protected]
Background: Despite improvements in diagnosis and treatment, lung cancer is one of the most lethal human diseases, with a dismal 5-year relative survival rate of only 5% for patients diagnosed with advanced metastatic disease. Accumulating evidence supports that epigenetic aberration of histone demethylase-KDM5 subfamily is linked to human pan-cancer. However, the detailed functions of KDM5 proteins in lung cancer, especially in non-small-cell lung cancer (NSCLC), remain poorly understand.
Methods: UALCAN, GEPIA, Kaplan–Meier plotter, cBioPortal, TIMER, TISIDB, and STRING databases were utilized in this investigation.
Results: We detected varying degrees of gene mutations of KDM5 subfamily members and found that KDM5B/C were remarkably overexpressed in LUAD and LUSC compared to normal tissues. Different from KDM5D, positive relationship was shown between overall survival and mRNA expression of KDM5A/B/C in lung cancer. We determined that KDM5A/B/C expression levels were positively correlated with CD4+ T cells infiltration, especially immunological markers of Tregs and Th17 cells. Moreover, LUAD and LUSC were separately rich in inflammatory and wound healing subtypes after immunogenomics analyzing with respect to KDM5 subfamily overexpression. And with their 120 co-expressed genes, we revealed that nucleocytoplasmic transport and cellular protein localization-related genes were closely connected to KDM5 subfamily alterations, next to chromatin remodeling genes.
Conclusion: We formulated the immune-infiltrating and prognostic value of KDM5 subfamily and highlighted its promising role in immune-inflammatory interaction with tumour microenvironment in NSCLC.
Keywords: non-small-cell lung cancer, biomarker, KDM5 subfamily, bioinformatics analysis, immune cell infiltration
Introduction
Lung cancer is one of the most lethal human diseases and the second common cancer expected to be diagnosed in men and women worldwide, with an overall 5-year survival rate of 19%.1 Despite improvements in early detection and treatment, the 5-year relative survival rate is only 5% for patients diagnosed with advanced metastatic disease (57%).2 There are two main forms of lung cancer: small cell lung cancer (SCLC, nearly 15% of the patients) and non-small-cell lung cancer (NSCLC, approximately 85% of the patients), with the latter further subdivided into 4 main types: lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), large cell carcinoma, and bronchial carcinoid tumour.3 Adenocarcinoma, that arises from alveolar cells located in smaller airway epithelium, is the most prevalent type of NSCLC and comprises of approximately 40% of lung cancer. Squamous cell carcinoma derives from cells located in the airway epithelium and represents 25–30% of lung cancer.4 Large cell cancer, typically poorly differentiated and composed of large cells with abundant cytoplasm and large nucleoli, accounting for 5–10% of all lung malignancy. And bronchial carcinoid can arise from main, lobar or segmental bronchi and accounting for approximately 0.5–2.5% of all lung cancer.3 In the past decade, incidence of NSCLC has declined due to advances in screening of precancerous lesions and systemic understanding of the molecular mechanisms of carcinogenesis. The advent of immune checkpoint inhibitors (ICIs) and several lines of tyrosine kinase inhibitors in cases with EGFR, ALK, ROS1, and NTRK mutations have dramatically changed the landscape of NSCLC therapy. However, deaths due to lung cancer are projected to increase prominently, a rate that exceeds the combined two next most common cancers (colorectal and prostate cancers) in 2020.1 Mortality data suggested that environmental exposures, endogenous hormones and sex differences co-worked in tumour microenvironment leading to a higher excess risk in male than female.1 Novel biomarkers are urgently needed and will most likely arise from a more elaborate understanding of tumour immune microenvironment and from the identification of genetic abnormalities in NSCLC.
Histone methylation is essential in regulating chromatin remodeling and gene transcriptional state, thereby its dysregulation has been observed in the developmental processes of various cancers.5 There are two classes of enzymes involved in maintaining its state: histone methyltransferases (HMTs) and histone demethylases (HDMs), such as lysine demethylase-KDM. Promoting the de-methylation of these repressive enzymes has a comparable effect as counteracting epigenetic gene regulation of HMTs. Other than KDM1 (also known as LSD), which removes methyl groups via flavin-dependent monoamine oxidation to regulate active chromatic state, multiple studies concentrated on KDM5 (also known as JARID1) enzymes, removing methyl groups through 2-oxoglutarate (2-OG)- and Fe(II)-dependent hydroxylation to modify multiple metabolic processes.6 The KDM5 subfamily is encoding a histone H3 lysine 4 demethylases enzyme that can function through both demethylase-dependent and -independent mechanisms to recognize and enzymatically regulate chromatin. Mammalian cells encode four KDM5 paralogs, KDM5A (JARID1A/RBP2), KDM5B (JARID1B/PLU-1), KDM5C (JARID1C/SMCX), and KDM5D (JARID1D/SMCY), whereas lower eukaryotes with smaller genomes each have a single ortholog.7 All four family members of KDM5 share sequence and structure similarity: a Jumonji N (JmjN) domain, a Jumonji C (JmjC) domain, a helical C5HC2 motif containing zinc finger (C5HC2-ZF) domain, a AT-rich interactive domain (ARID), and two or three plant homeodomains (PHDs).8 The ARID and PHD domains separate the catalytic domain into two fragments (JmjN and JmjC), and the JmjC catalytic domain demethylates histone H3 trimethylated at lysine 4 is unique to affect transcriptional repression of KDM5 proteins.9 KDM5A and KDM5B have a third PHD domain (PHD3) and are involved in tumour suppression by enhancing cellular senescence.7 Meanwhile, KDM5C and KDM5D are implicated in later tissue homeostasis and located on the X and Y chromosomes, respectively.10,11 A substantial study identifies that KDM5 demethylases are implicated in various biological and pathological processes including the negative regulation of tumorigenesis.12 In addition to poising a series of epigenetic events, a further study revealed their roles in mediating drug tolerance after cancer therapy.13
Although research lends support to the notion that KDM5 subfamily as a potential candidate for epigenetic-based antitumor therapy in human pan-cancers,14–17 the detailed functions of KDM5 proteins in lung cancer, especially in non-small-cell lung cancer (NSCLC), remain uncertain. In this context, our research will focus on the enzymatic activity of KDM5 and its immunological interaction with tumour immune infiltration in NSCLC. Our findings mapped KDM5 demethylases as attractive biomarkers to govern immune cell recruitment and infiltration in NSCLC.
Materials and Methods
Ethics Statement
Since all the data were retrieved from the online databases, it could be affirmed that all written informed consents had already been obtained.
UALCAN
UALCAN is a tool for facilitating tumour subgroup gene expression and survival analyses based on The Cancer Genome Atlas (TCGA), which contained clinical data from 31 cancer types (http://ualcan.path.uab.edu). In our study, UALCAN was used to illustrate the distinct expression levels of KDM5A subfamily members in lung tumour and adjacent normal tissues. Student’s t-test was used to generate a p-value and the p-value cutoff was 0.05.
GEPIA
GEPIA provides customizable functions such as tumour/normal differential expression analysis, profiling according to cancer types or pathological stages, patient survival analysis, similar gene detection, and correlation analysis of 9736 tumours and 8587 normal samples from the TCGA and the GTEx projects (http://gepia.cancer-pku.cn/). The Student’s t-test was used to generate a p-value and the p-value cutoff was 0.05.
TIMER
TIMER (Tumor IMmune Estimation Resource) provides systematical analysis of immune infiltrations estimated by multiple immune deconvolution methods across diverse cancer types (https://cistrome.shinyapps.io/timer/). In our study, TIMER was used to generate high-quality figures dynamically to explore KDM5-related tumour immunological, clinical and genomic features comprehensively.
TISIB
TISIDB is a portal for elucidating the interaction between tumour and immune system to assist predicting immunotherapy responses in tumour microenvironment according to PubMed and TCGA databases (http://cis.hku.hk/TISIDB/). Further to develop novel immunotherapy target that play a crucial role in cancer initiation, progression, and treatment.
Kaplan–Meier Plotter
The Kaplan–Meier plotter is capable of assessing the effect of 54k genes to provide meta-analysis in line with discovery and validation of survival biomarkers across 21 cancer types (https://kmplot.com/analysis/). In this study, it was used to evaluate the prognostic value of KDM5 subfamily mRNA expression in which the lung cancer patients were split into high and low expression groups based on median values of mRNA expression and validated by progression-free survival (FP), post-progression survival (PPS), and overall survival (OS) curves, with the hazard ratio (HR) of 95% confidence intervals (CI) and logrank p-value. The statically significant difference was considered when a p-value is <0.05.
TCGA Data and cBioPortal
The Cancer Genome Atlas (TCGA), a landmark cancer genomics program, is molecularly characterized over 20,000 primary cancers and matched normal samples spanning 33 cancer types. The LUAD (TCGA, Firehose Legacy; 586 cases) and LUSC (TCGA, Firehose Legacy; 511 cases) datasets including data with pathology reports of KDM5 subfamily were selected for further analyses using the cBioPortal for Cancer Genomics, which is a comprehensive portal for exploring, visualizing, and analyzing multidimensional cancer genomics database (http://www.cbioportal.org/). In this study, we analyzed the genomic profiles of KDM5A/B/C/D members, which contained mutations, putative copy-number alterations, and mRNA Expression Scores. Co-expressed genes of KDM5 subfamily were performed with the “Co-expression” module of cBioportal. Pearson’s correlation coefficient was used to investigate the correlation between KDM5 subfamily and co-expressed genes and the top co-expressed genes of KDM5A/B/C subfamily with the largest Pearson’s correlation coefficient were listed.
STRING
The protein–protein interaction (PPI) network was retrieved from Search Tool for the Retrieval of Interaction Gene/Proteins (STRING) (https://string-db.org/) database with high confidence. The interactions include direct (physical) and indirect (functional) associations; they stem from computational prediction, from knowledge transfer between organisms, and from interactions aggregated from other (primary) databases. In STRING, each protein–protein interaction is annotated with one or more ‘scores’. All scores rank from 0 to 1, with 1 being the highest possible confidence. A score of 0.5 would indicate that roughly. In this study, STRING was used to map the top 120 co-expressed genes of KDM5A/B/C subfamily.
Results
Aberrant Expression of KDM5 Subfamily in Patients with NSCLC
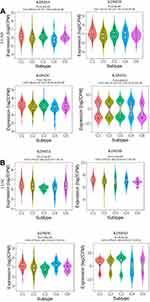
The KDM5 subfamily genes are characterized by having a JmjC domain, which encoded histone demethylase activity. Loss and overexpression of KDM5 proteins are linked to intellectual disability and cancer, respectively. GEPIA was utilized to explore differential transcription levels of KDM5A/B/C/D between tumour and adjacent normal tissues in LUAD and LUSC based on TCGA database. Distributions of gene expression levels are displayed using box plots, with statistical significance of differential expression evaluated using the Wilcoxon test. We can identify genes that are up- or down-regulated in tumours compared to normal tissues for each cancer type, as displayed in gray columns when normal data are available. As shown in Figure 1, the expression levels of KDM5B/C were remarkably higher in LUAD and LUSC than normal tissues. What is more, significant overexpression of KDM5A was found in LUSC tissues compared to normal tissues. However, the difference of KDM5D comparative was not obvious. Furthermore, using the UALCAN, we compared the mRNA expression of KDM5 subfamily between 515 LUAD and 59 normal tissues, 503 LUSC and 52 normal tissues, respectively. The results in Figure 2A and B revealed an absolutely increase of KDM5B/C mRNA expression in LUAD and LUSC tissues. The relative level of KDM5A was prominently higher in LUSC than in lung tissues, although it was undifferentiated between LUAD and normal lung tissues. Besides, we also contrast the relative expression levels of KDM5 subfamily members in LUAD and LUSC tissues (GEPIA) and determined that among all the factors we evaluated, KDM5C was the highest expression in both LUAD and LUSC (Figure 2C). Taken together, our results showed that KDM5A/B/C were significant up-regulated in patients with LUSC, whereas KDM5B/C were obviously over-expressed in patients with LUAD. The reason why the mRNA expression of KDM5 subfamily seemed to significantly diverge may be related to patients’ individual cancer pathology and as a result of small sample size.
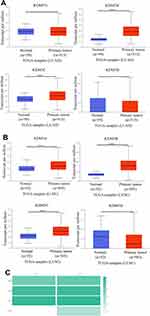 |
Figure 2 The mRNA expression of KDM5 subfamily members between 515 LUAD and 59 normal tissues (A), 503 LUSC and 52 normal tissues (B). ***p<0.001. The relative level of KDM5 subfamily in NSCLC (C). |
Genetic Mutations and Prognostic Features of KDM5 Subfamily in Patients with Lung Cancer
Epigenetic aberrations play an important role in early malignancies. Here, we analyzed the KDM5 subfamily gene alterations and correlations by using cBioPortal online tool for LUAD (TCGA, Firehose Legacy), in which KDM5 subfamily was varied in 517 samples out of 586 patients (Figure 3A). Gene alterations in KDM5B (25%) were most frequently observed, followed by mutations in KDM5A (15%), KDM5C (8%), and KDM5D (0.4%). Simultaneously, we investigated the correlations between KDM5 subfamily gene alterations and LUSC (TCGA, Firehose Legacy), in which the genes were varied in 501 samples out of 511 patients (Figure 3B). And KDM5A was the top genetic alterations with a rate of 25%.
By means of data mining in Kaplan–Meier plotter, prognostic value of KDM5 subfamily for lung cancer patients including FP, PPS, and OS were separately figured. It could be seen that, in each cohort, patients were divided into low- and high-risk groups based on cutoff value (Figure 3C–F). Reversed relationship was shown between OS and the mRNA level of KDM5D; however, the low expression groups of KDM5A/B/C were shown shorter OS in lung cancer. In addition, decreased KDM5A or KDM5B mRNA expression led to a reduced PPS. There was no significant correlation between FP and KDM5B. This discovery observed that the changes of KDM5 subfamily genes might affect prognosis of NSCLC patients.
Correlation Between KDM5 Subfamily Expression and Abundance of Immune Cell Infiltration in NSCLC
The biological process of inflammation relies on diverse immune cell type infiltration. Immune cells are an important component that can secrete inflammatory mediators to affect tumour microenvironment.18 Subsequently, we explored the relationship between KDM5A/B/C/D expression and the degree of immune cell infiltration using the TIMER database. The abundances of six immune infiltrates (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) were analyzed in LUAD and LUSC (Figure 4). Scatterplots showed the purity-corrected partial Spearman’s rho value and statistical significance. KDM5A expression was positively associated with levels of CD8+ T cells (R = 0.252, P = 1.76e-08), CD4+ T cells (R = 0.222, P = 7.78e‐07), neutrophils (R = 0.332, P = 7.30e‐14), macrophages (R = 0.201, P = 8.47e‐06), and Dendritic cells (R = 0.218, P = 1.14e‐06) in LUAD, whereas it was significantly associated with levels of CD4+ T cells (R = 0.18, P = 7.82e-05) in LUSC. In addition to CD4+ T cells, no significant correlations between KDM5B/C expression and infiltrating levels of other immune cells were observed. Unexpectedly, there was no such relationship between KDM5D expression and immune cell infiltration in NSCLC.
Furthermore, somatic copy number alterations (SCNAs), including deep deletion, arm-level deletion, diploid/normal, arm-level gain, and high amplification, were applied to show distribution of each immune subset at each copy number status in NSCLC with KDM5 subfamily mutations (Supplementary Figure 1). The infiltration level for each SCNA category was compared with the normal using a two-sided Wilcoxon rank-sum test. Most of the copy number varies distributed in arm-level gain of both LUAD and LUSC. Unfortunately, by querying this public database, we did not find the relationship between KDM5D and SCNAs.
Assessment of the Relationship Between KDM5A/B/C and Immune Marker Expression
We further explored the relationship between KDM5A/B/C expression and levels of immune cell infiltration based on a variety of immunological markers in LUAD and LUSC using the TIMER and GEPIA databases (Table 1). The immune markers for particular cell subsets including CD8+ T cells, total T cells, B cells, monocytes, tumour-associated macrophages (TAMs), M1 and M2 macrophages, neutrophils, natural killer cells (NK cells), dendritic cells (DCs), T helper cells (Th1, Th2, Th17 cells), Tfh cells (follicular helper T cells), regulatory T cells (Tregs) and exhausted T cells. We adjusted these results based on tumour purity, revealing a significant correlation between KDM5A expression and monocyte markers (CD86, CD115), TAM marker (CD68), M1 macrophage markers (INOS, COX2), M2 macrophage marker (CD163), neutrophils marker (CD11b), NK cell marker (KIR2DL3), DC markers (BDCA-4, CD11C), Th1 markers (T-bet, STAT1), Th2 markers (GATA3, STAT6, STAT5A), Tfh markers (BCL6, IL21), Th17 marker (STAT3) and Treg markers (FOXP3, CCR8, STAT5B, TGFβ1) in LUAD. A significant correlation between KDM5B expression and total T cell marker (CD3D), M1 macrophage marker (COX2), Th1 marker (STAT1), Th2 marker (STAT6), Tfh marker (BCL6), Th17 marker (STAT3) and Treg marker (STAT5B) in LUAD. Elevated KDM5C expression was related with M1 macrophage marker (IRF5), DC markers (BDCA-4, CD11c), Th1 marker (TNF), Th2 markers (STAT6, STAT5A), Tfh marker (BCL6), Th17 marker (STAT3) and Treg markers (FOXP3, CCR8, STAT5B) in LUAD.
 |
 |
 |
Table 1 Correlation Analysis Between KDM5A/B/C and Markers of Immune-Infiltrating Cells |
In LUSC, KDM5A expression was associated with monocyte marker (CD115), M1 macrophage marker (INOS), neutrophil marker (CD11b), DC marker (BDCA-4), Th1 markers (STAT4, STAT1), Th2 markers (GATA3, STAT6, STAT5A), Tfh marker (BCL6), Th17 marker (STAT3) and Treg markers (FOXP3, CCR8, STAT5B). CD8+T cell marker (CD8A), total T cell markers (CD3D, CD2), M1 macrophage marker (COX2), Th1 marker (IFNG), Th2 markers (GATA3, STAT6), Tfh marker (BCL6), Th17 marker (STAT3), Treg marker (STAT5B) and T cell exhaustion markers (TIM-3, GZMB) were involved in KDM5B expression. And levels of M1 macrophage marker (INOS), neutrophil markers (CD11b, CCR7), DC marker (CD11c), Th1 marker (T-bet), Th2 markers (STAT6, STAT5A), Tfh marker (BCL6), Th17 marker (STAT3), Treg marker (STAT5B) were relatively high with KDM5C overexpression. There was almost no association between KDM5D and immune marker expression in neither LUAD nor LUSC.
We therefore observed that levels of KDM5A/B/C were correlated with the majority of Th17 and Treg markers (Figure 5), indicating their complicate roles in immune cell recruitment of NSCLC, and further work will be needed to explore the mechanisms underlying such immune infiltration in tumour microenvironment.
 |
Figure 5 Scatterplots of correlation analysis between KDM5A/B/C/D and immunological markers of Treg (STAT5B) (B) and Th17 (STAT3) (A) cells in LUAD and LUSC using TIMER. |
The Immune Landscape of KDM5 Subfamily Expression in NSCLC
Thorsson et al identified six immune subtypes: wound healing, IFN-γ dominant, inflammatory, lymphocyte depleted, immunologically quiet, and TGF-β dominant, based on immunogenomic analyses of more than 10,000 tumors. They are different in lymphocyte signatures, intratumoral heterogeneity, aneuploidy, neoantigen load, overall cell proliferation, immunomodulatory genes, and prognosis.19 As shown in Figure 6, LUAD was rich in C3 (inflammatory) subtype associated with KDM5A/B/C, which was defined by elevated Th17 and Th1 genes in line with TIMER database. It is characterized with low to moderate tumour cell proliferation, lower levels of aneuploidy and overall somatic copy number alterations compared with other subtypes.19 In LUSC, overexpression levels of KDM5A/B/C were rich in C1 (wound healing) subtype, which had elevated expression of angiogenic genes, a high proliferation rate, and a Th2 cell bias to adaptive immune infiltrate.19
Intriguingly, different pathologic category of lung cancer associated with KDM5 subfamily overexpression was not rich in C5 (immunologically quiet) subtype, which exhibited the lowest lymphocyte and highest macrophage responses, dominated by M2 macrophages.
Gene Enrichment Analysis of KDM5 Subfamily and Their 120 Co-Expression Genes in NSCLC Patients
After analyzing the KDM5 subfamily genetic alterations, correlation with immune cell infiltration, and their prognostic value in LUAD and LUSC patients, we next analyzed 120 co-expression genes (KDM5A/B/C related) that were significantly associated with KDM5 subfamily mutations and listed them in Supplementary Table 1. Subsequently, protein–protein interaction network was constructed by STRING (Supplementary Figure 2). Gene enrichment analysis was employed to analyze their 120 co-expression genes. As shown in Figure 7A, we found that GO:0016569 (covalent chromatin modification), GO:0006338 (chromatin remodeling), CORUM:6470 (Set1B complex), GO:0071557 (histone H3-27 demethylation), and GO:006913 (nucleocytoplasmic transport) were remarkably correlated with KDM5A/B/C mutations in LUAD, whereas GO:0016569 (covalent chromatin modification), GO:1902275 (regulation of chromatin organization), GO:0006338 (chromatin remodeling), GO:1903827 (regulation of cellular protein localization), and hsa04919 (thyroid hormone signaling pathway) were predominantly associated with the KDM5 subfamily alterations in LUSC (Figure 7B). Altogether, the results revealed that nucleocytoplasmic transport-related genes, such as YAP1 and TAOK1, were closely connected with KDM5 subfamily alterations in NSCLC.
 |
Figure 7 Gene enrichment analysis of KDM5A/B/C subfamily members and their 120 co-expression genes in LUAD (A) and LUSC (B) patients. |
Discussion
There is a growing understanding of the oncogenic and tumour suppressive functions of histone demethylase family, which regulates a subset of transcribed genes at promoters and participates in chromatin remodeling.20 KDM5 demethylase subfamily contains multiple conserved domains and facilitates the removal of methyl group on di- and trimethylated forth lysine of histone H3 (H3K4).21 The isoforms contain specific cosubstrates, such as oxygen, α-KG, and vitamin C, making them potential players in tumorigenesis, transcriptional plasticity, and epigenetic alterations contingent on cancer cellular context.5 Dysregulation of KDM5 subfamily genes destroys the balance between “writer” and “eraser” enzymes leading to defects in histone posttranslational modifications involving multiple human disorders. Unlike other histone modification enzymes, KDM5 could recognize specific DNA sequences in their targets owing to the ARID domain.9 Despite their high homology, members of the KDM5 subfamily differ in physiological and pathological functions.22 Furthermore, the roles of KDM5 demethylases in regulating normal cell functionality and altering tumour microenvironment remain ill-defined in lung cancer, especially in NSCLC. Therefore, the present study is the first time to analyze the mRNA expression, gene alterations, immune cell infiltration, and prognostic role of KDM5 subfamily in NSCLC.
Studies indicated that KDM5A was associated with tumour suppression by mediating RB interacting proteins via repressing E2F target genes.23,24 However, knockdown of KDM5A altered H3K4 methylation suppressing tumour proliferation, which suggested its oncogenic role.25 KDM5A overexpression has been observed in glioblastoma, gastric and hepatocellular carcinoma26–28 and it is recognized to increase levels of cyclin-dependent kinase inhibitors p21 and p27 through senescence pathways in lung tumorigenesis.29 With TCGA data set analysis, we assessed the clear difference between tumour and adjacent normal tissues expression levels of KDM5A in LUSC, although there was no significance of KDM5A expression in LUAD. What is more, the mRNA level of KDM5A was found to be noticeably over-expressed and gene alterations were most frequently observed (25%) in LUSC. Altered KDM5A expression between LUAD and LUSC may be due to differences in the underlying biological mechanisms.
KDM5B was identified as a tumour suppressor and upregulated by overexpression of c-ErbB2.30 It has been observed to associate with cell cycle, drug tolerance, and oxidative metabolism and overexpressed KDM5B has also been described associated with upregulated E2F/RB pathway in lung cancer.31 Specifically, TCGA predicted KDM5C mutation as a cancer driver, although controversially identified as one of the few genes on the X chromosome that exhibit oncogenic and anti-oncogenic properties.32 It has been revealed that KDM5C mediate miR-133a expression to suppress development and metastases of lung cancer by targeting PTBP1.33 Across these databases, we consistently observed elevated expression levels of KDM5B/C and relatively high gene mutations in either LUAD or LUSC, which remind us of their oncogenic properties under cancer type-specific context.
KDM5D, recognized as a sexually dimorphic gene, is 85% identical to KDM5C and more frequently encoded in the Y chromosome of all male tissues.34,35 Here, we deciphered that KDM5D gene mutations were prominently lower than other KDM5 members in NSCLC. Higher KDM5D expression led to a reduced FP and OS in LUAD and LUSC patients, regardless of the lymphatic metastasis status. There was a particularly strong correlation between high KDM5 subfamily expression and poor gastric cancer prognosis using Kaplan–Meier plotter. Noticeably, low KDM5A/B/C expression correlated with reduced OS, indicating different potential prognosis signatures of KDM5A/B/C/D in NSCLC.
An additional key finding in this investigation was that KDM5A/B/C expression correlated with the degree of immune cell infiltration in NSCLC. Immune system is recognized as a complex endeavor that eradicates foreign antigens in non-malignant setting, versus hijacked by tumour-specific antigens in progression and metastasis stages.36 In addition to serving as the first line of defense against multiple pathogens, immune cells provide surveillance via identifying and destroying occult cancerous cells, and contrary aid tumour to evade immune control under certain circumstances.37 Tumour-infiltrating lymphocytes are a special group of lymphocytes that infiltrate the tumour microenvironment by detecting cancer antigens and releasing pro-inflammatory cytokines.38
In this investigation, the expression levels of KDM5A/B/C were positively correlated with CD4+ T cells infiltration, especially the degree of Tregs and Th17 cells, and negatively correlated with natural killer cell infiltration (Table 1). CD4+ T cells differentiate into distinct Th cells, including Th1, Th2, Th17, Treg, Th9, Th22, and T follicular helper cells, exert divergent functions depending on tumour-secreted cytokines.39,40 Th17 cells, characterized by secreting IL-17A, IL-17F, IL-21, IL-22, and CCL20, expressing master transcription factor RORγt, and promoting inflammation in response to infections, have complicated roles in carcinogenesis.41 Tregs are characterized by the expression of inhibitory cytokines such as IL-10 and TGF-β or mediate inhibitory checkpoint molecules such as TIGIT and CTLA-4 to suppress immune response and promote tumorigenesis.42
Generally, the relevance of Th17 cells have been documented in promoting autoimmunity, whereas Treg cells are critical for immune tolerance and have been shown to dampen autoimmunity and antitumor immunity.43,44 Over the past decade, a major focus has been laid on Th17/Tregs dynamics in tweaking the immune system of lung cancer.45 Patients were found to have a greater ratio of Th17/Treg frequencies compared to healthy controls, which correlated with the levels of carcinoembryonic antigen (CEA) in non-small cell lung cancer.45 A small fraction of studies indicated greater numbers of Th17 cells negatively correlated with overall survival in lung cancer tissues, and IL-17 neutralization reduced metastatic nodules in mice.46 Additionally, a novel mechanism revealed Th17 differentiated from Tregs via IL-2 to promote inflammation and tumorigenesis in colon cancer.47 Herein, more studies are needed to figure out the relationship between KDM5A/B/C overexpression and Th17/Tregs in the context of the lung tumour microenvironment.
Immune subtypes were employed to provide potential therapeutic and prognostic implications for lung cancer management. LUAD and LUSC associated with overexpressed KDM5A/B/C were separately rich in inflammatory and wound healing subtypes. Accordingly, inflammatory subtype demonstrated the most pronounced Th17 signature and the best prognosis, while wound healing subtype had less favorable outcomes despite having a substantial immune component,19 reflecting complex associations between immune response and differed tumour types.
We further analyzed the gene enrichment of KDM5 subfamily alterations in LUAD and LUSC respectively. The results in Figure 7A exposed that nucleocytoplasmic transport and cellular protein localization-related genes, such as YAP1 and TAOK1, were closely connected with KDM5 family alterations, next to chromatin remodeling genes. However, further study will be needed to confirm the mechanisms underlying such relationship. Currently, KDM5 subfamily could be recognized as a promising biomarker for prognosis and immune cell infiltration in lung cancer according to our investigation. The molecular mechanisms underlying Th17/Tregs recruitment and pro-inflammatory/immunosuppressive functions correlated with KDM5A/B/C in the lung tumour microenvironment require further study.
Conclusion
In summary, we formulated the immune-infiltrating and prognostic value of KDM5 subfamily from comprehensively analyzing its relationship with survival, gene alterations, and tumour immune infiltration cells in NSCLC. After evaluating a variety of markers associated with particular immune cell subsets, we recognized the pronounced role of KDM5 subfamily in immune cell recruitment. However, sequenced experiments and prospective clinical trials of its effect remain absent.
Data Sharing Statement
All data generated or analyzed during this study are included in this submitted article.
Ethics Approval and Consent to Participate
The study received approval from the institutional review board of Tianjin Medical University General Hospital.
Funding
There is no funding to report.
Disclosure
The author reports no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Toumazis I, Bastani M, Han SS, Plevritis SK. Risk-based lung cancer screening: a systematic review. Lung Cancer. 2020;147:154–186. doi:10.1016/j.lungcan.2020.07.007
3. Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. Introduction to the 2015 World Health Organization classification of tumors of the lung, pleura, thymus, and heart. J Thorac Oncol. 2015;10(9):1240–1242. doi:10.1097/JTO.0000000000000663
4. Takamochi K, Ohmiya H, Itoh M, et al. Novel biomarkers that assist in accurate discrimination of squamous cell carcinoma from adenocarcinoma of the lung. BMC Cancer. 2016;16(1):760. doi:10.1186/s12885-016-2792-1
5. Højfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov. 2013;12(12):917–930. doi:10.1038/nrd4154
6. Arifuzzaman S, Khatun MR, Khatun R. Emerging of lysine demethylases (KDMs): from pathophysiological insights to novel therapeutic opportunities. Biomed Pharmacother. 2020;129:110392.
7. Blair LP, Cao J, Zou MR, Sayegh J, Yan Q. Epigenetic regulation by lysine demethylase 5 (KDM5) enzymes in cancer. Cancers. 2011;3(1):1383–1404. doi:10.3390/cancers3011383
8. Dorosz J, Kristensen LH, Aduri NG, et al. Molecular architecture of the Jumonji C family histone demethylase KDM5B. Sci Rep. 2019;9(1):4019. doi:10.1038/s41598-019-40573-y
9. Pilka ES, James T, Lisztwan JH. Structural definitions of Jumonji family demethylase selectivity. Drug Discov Today. 2015;20(6):743–749. doi:10.1016/j.drudis.2014.12.013
10. Rondinelli B, Schwerer H, Antonini E, et al. H3K4me3 demethylation by the histone demethylase KDM5C/JARID1C promotes DNA replication origin firing. Nucleic Acids Res. 2015;43(5):2560–2574. doi:10.1093/nar/gkv090
11. Li N, Dhar SS, Chen TY, et al. JARID1D is a suppressor and prognostic marker of prostate cancer invasion and metastasis. Cancer Res. 2016;76(4):831–843. doi:10.1158/0008-5472.CAN-15-0906
12. Plch J, Hrabeta J, Eckschlager T. KDM5 demethylases and their role in cancer cell chemoresistance. Int J Cancer. 2019;144(2):221–231. doi:10.1002/ijc.31881
13. Hinohara K, Wu HJ, Vigneau S, et al. KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell. 2018;34(6):939–953.e9. doi:10.1016/j.ccell.2018.10.014
14. Oser MG, Sabet AH, Gao W, et al. The KDM5A/RBP2 histone demethylase represses NOTCH signaling to sustain neuroendocrine differentiation and promote small cell lung cancer tumorigenesis. Genes Dev. 2019;33(23–24):1718–1738. doi:10.1101/gad.328336.119
15. Bamodu OA, Huang W-C, Lee W-H, et al. Aberrant KDM5B expression promotes aggressive breast cancer through MALAT1 overexpression and downregulation of hsa-miR-448. BMC Cancer. 2016;16(1):160. doi:10.1186/s12885-016-2108-5
16. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49.
17. Komura K, Jeong SH, Hinohara K, et al. Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression. Proc Natl Acad Sci. 2016;113(22):6259–6264. doi:10.1073/pnas.1600420113
18. Kaewkangsadan V, Verma C, Eremin MJ, Cowley G, Ilyas M, Eremin O. Tumour-draining axillary lymph nodes in patients with large and locally advanced breast cancers undergoing neoadjuvant chemotherapy (NAC): the crucial contribution of immune cells (effector, regulatory) and cytokines (Th1, Th2) to immune-mediated tumour cell death induced by NAC. BMC Cancer. 2018;18:123.
19. Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity. 2018;48:812–830.
20. Gong F, Miller KM. Histone methylation and the DNA damage response. Mutat Res. 2019;780:37–47. doi:10.1016/j.mrrev.2017.09.003
21. Lee DH, Kim GW, Jeon YH, Yoo J, Lee SW, Kwon SH. Advances in histone demethylase KDM4 as cancer therapeutic targets. FASEB J. 2020;34(3):3461–3484. doi:10.1096/fj.201902584R
22. Chi P, Allis CD, Wang GG. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10(7):457–469. doi:10.1038/nrc2876
23. Chicas A, Kapoor A, Wang X, et al. H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proc Natl Acad Sci USA. 2012;109(23):8971–8976. doi:10.1073/pnas.1119836109
24. Defeo-Jones D, Huang PS, Jones RE, et al. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature. 1991;352(6332):251–254. doi:10.1038/352251a0
25. Hou J, Wu J, Dombkowski A, et al. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am J Transl Res. 2012;4(3):247–256.
26. Romani M, Daga A, Forlani A, Pistillo MP, Banelli B. Targeting of histone demethylases KDM5A and KDM6B inhibits the proliferation of temozolomide-resistant glioblastoma cells. Cancers. 2019;11(6):878. doi:10.3390/cancers11060878
27. Zeng J, Ge Z, Wang L, et al. The histone demethylase RBP2 is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology. 2010;138(3):981–992. doi:10.1053/j.gastro.2009.10.004
28. Liang X, Zeng J, Wang L, et al. Histone demethylase retinoblastoma binding protein 2 is overexpressed in hepatocellular carcinoma and negatively regulated by hsa-miR-212. PLoS One. 2013;8(7):e69784. doi:10.1371/journal.pone.0069784
29. Teng YC, Lee CF, Li YS, et al. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer Res. 2013;73(15):4711–4721. doi:10.1158/0008-5472.CAN-12-3165
30. Lu PJ, Sundquist K, Baeckstrom D, et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem. 1999;274(22):15633–15645. doi:10.1074/jbc.274.22.15633
31. Roesch A, Vultur A, Bogeski I, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23(6):811–825. doi:10.1016/j.ccr.2013.05.003
32. Niu X, Zhang T, Liao L, et al. The von Hippel–Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C. Oncogene. 2012;31(6):776–786. doi:10.1038/onc.2011.266
33. Zhang Q, Xu L, Wang J, et al. KDM5C expedites lung cancer growth and metastasis through epigenetic regulation of microRNA-133a. Onco Targets Ther. 2021;14:1187–1204. doi:10.2147/OTT.S288799
34. Akimoto C, Kitagawa H, Matsumoto T, Kato S. Spermatogenesis-specific association of SMCY and MSH5. Genes Cells. 2008;13(6):623–633. doi:10.1111/j.1365-2443.2008.01193.x
35. Willis-Owen SAG, Domingo-Sabugo C, Starren E, et al. Y disruption, autosomal hypomethylation and poor male lung cancer survival. Sci Rep. 2021;11(1):1245. doi:10.1038/s41598-021-91907-8
36. Saab S, Zalzale H, Rahal Z, Khalifeh Y, Sinjab A, Kadara H. Insights into lung cancer immune-based biology, prevention, and treatment. Front Immunol. 2020;11:159. doi:10.3389/fimmu.2020.00159
37. Shalapour S, Karin M. Immunity, inflammation, and cancer: an eternal fight between good and evil. J Clin Invest. 2015;125(9):3347–3355. doi:10.1172/JCI80007
38. Lee N, Zakka LR, Mihm MC
39. Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28(1):445–489. doi:10.1146/annurev-immunol-030409-101212
40. Kim HJ, Cantor H. CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol Res. 2014;2(2):91–98. doi:10.1158/2326-6066.CIR-13-0216
41. Chang SH. T helper 17 (Th17) cells and interleukin-17 (IL-17) in cancer. Arch Pharm Res. 2019;42(7):549–559. doi:10.1007/s12272-019-01146-9
42. Moreno Ayala MA, Li Z, DuPage M. Treg programming and therapeutic reprogramming in cancer. Immunology. 2019;157(3):198–209. doi:10.1111/imm.13058
43. Tanaka A, Sakaguchi S. Targeting treg cells in cancer immunotherapy. Eur J Immunol. 2019;49(8):1140–1146. doi:10.1002/eji.201847659
44. Wing JB, Tanaka A, Sakaguchi S. Human FOXP3(+) regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity. 2019;50(2):302–316. doi:10.1016/j.immuni.2019.01.020
45. Duan MC, Han W, Jin PW, et al. Disturbed Th17/Treg balance in patients with non-small cell lung cancer. Inflammation. 2015;38(6):2156–2165. doi:10.1007/s10753-015-0198-x
46. Salazar Y, Zheng X, Brunn D, et al. Microenvironmental Th9 and Th17 lymphocytes induce metastatic spreading in lung cancer. J Clin Invest. 2020;130(7):3560–3575. doi:10.1172/JCI124037
47. Geis AL, Fan H, Wu X, et al. Regulatory T-cell response to enterotoxigenic bacteroides fragilis colonization triggers IL17-dependent colon carcinogenesis. Cancer Discov. 2015;5(10):1098–1109. doi:10.1158/2159-8290.CD-15-0447
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.