Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
Systematic Pan-Cancer Analysis of KIF23 and a Prediction Model Based on KIF23 in Clear Cell Renal Cell Carcinoma (ccRCC)
Authors Bai X , Cao Y, Yan X, Tuoheti K, Du G, Chen Z, Wu H, Guo L, Liu T
Received 10 September 2021
Accepted for publication 2 December 2021
Published 30 December 2021 Volume 2021:14 Pages 1717—1729
DOI https://doi.org/10.2147/PGPM.S337695
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Xiaojie Bai, Yuanfei Cao, Xin Yan, Kurerban Tuoheti, Guowei Du, Zhao Chen, Huahui Wu, Linfa Guo, Tongzu Liu
Department of Urology, Zhongnan Hospital of Wuhan University, Wuhan, People’s Republic of China
Correspondence: Tongzu Liu
Department of Urology, Zhongnan Hospital of Wuhan University, 169 Donghu Road, Wuhan, 430071, People’s Republic of China
Tel +86-27-67813104
Fax +86-27-67812667
Email [email protected]
Purpose: This study aims to carry out a pan-cancer analysis of kinesin family member 23 (KIF23) and construct a predictive model for the prognosis of clear cell renal cell carcinoma (ccRCC) patients.
Methods: We evaluated the differential expression of KIF23 in pan-cancer by The Cancer Genome Atlas (TCGA) and Oncomine database. Then, the correlation between KIF23 with prognosis, clinical grade, stage, immune subtype, tumor mutation burden (TMB), microsatellite instability (MSI) and immune microenvironment was explored by TCGA, an integrated repository portal for tumor-immune system interactions (TISIDB) and cBioPortal. Subsequently, we screened out ferroptosis-related genes (FRGs) related to KIF23 and constructed a risk score model. Univariate Cox analysis was used to determine independent prognostic factors for ccRCC overall survival (OS), and a nomogram was established. Furthermore, gene set enrichment analysis (GSEA) was applied to study the biological functions and pathways of KIF23. Finally, quantitative real-time polymerase chain reaction (qRT-PCR) was carried out to evaluate the expression of KIF23.
Results: KIF23 was highly expressed in most tumors. Further, KIF23 was strongly correlated with prognosis, clinical grade, stage, immune subtype, TMB, MSI and immune microenvironment in different tumors. We found that KIF23 was significantly associated with all aspects of ccRCC. Then, 8 FRGs were identified to construct a risk score model together with KIF23. And a prognostic nomogram prediction model of OS was established. After GSEA analysis, cell cycle, condensed chromosome and other physiological processes were screened out. Finally, qRT-PCR verified the high expression of KIF23 in ccRCC cell lines than normal kidney cell line.
Conclusion: KIF23 may act as a pivotal part in occurrence and progression of different tumors. In ccRCC, KIF23 can be a great prognostic biomarker, and the nomogram based on KIF23 may contribute to better treatment plans for ccRCC patients.
Keywords: clear cell renal cell carcinoma, ferroptosis, nomogram
Introduction
According to the latest Global Cancer Statistics, the incidence of renal cell carcinoma (RCC) ranks 12th, with approximately 74,000 new cases in the United States in 2020.1 Clear cell renal cell carcinoma (ccRCC) accounts for 70–80% of all RCC and is one of the most aggressive pathological type.2 For localized ccRCC, surgical interventions are the best options.3 However, about 30% of renal cancer patients already present with metastasis at the time of initial diagnosis since there are no obvious symptoms in the early stage.4 One quarter of surgically treated patients will eventually develop cancer recurrence and metastasis.5 Although treatment options have advanced a lot, the prognosis of this disease remains poor, especially for patients with advanced stages.6 Therefore, this calls for exploration of more effective prognostic biomarkers and construction of more accurate prognostic models to provide better treatment decisions.
Kinesin family member 23 (KIF23), also called MKLP1, is the human homolog of mouse KIF23. Previous research indicated that KIF23 played an important role in regulation of cytokinesis.7 Dysfunction of KIF23 led to mitotic arrest and the generation of binucleated or multinucleated cells, resulting in the appearance of tumors.8 Previous studies have also shown that KIF23 was highly expressed in a variety of tumors and was correlated with cancer progression, such as gastric cancer,9 gliomas10 and breast cancer.11 Knockdown of KIF23 suppressed proliferation of pancreatic ductal adenocarcinoma cells.12 These studies suggested that KIF23 may be an ideal prognostic and therapeutic pan-cancer biomarker.
Ferroptosis is a novel form of iron-dependent regulated cell death, which is different from apoptosis and autophagy. The key characteristic of Ferroptosis is the accumulation of excessive reactive oxygen species (ROS) and abnormal iron metabolism. Increasing evidence has shown that Ferroptosis is involved in the progression and metastasis of cancers. Meanwhile, Ferroptosis has also became a potential target for cancer therapy.
In this study, we utilized the TCGA database and Oncomine to evaluate the differential expression and the prognosis value of KIF23 gene in pan-cancer. We also explored the correlation of KIF23 expression with clinicopathological features (tumor stage and grade) and immune subtype based on TISIDB. Meanwhile, genomic alteration of KIF23 was displayed by using cBioPortal. The correlation between KIF23 and microsatellite instability (MSI), tumor mutational burden (TMB) and tumor microenvironment was studied too. Then we screened out FRGs related to KIF23 and constructed a prognosis prediction model for ccRCC patients by combining lasso regression and cox regression analysis. GSE29609 was employed to validate this model. Furthermore, gene set enrichment analysis (GSEA) was applied to study the biological functions and pathways of KIF23. Finally, we performed qRT-PCR to verify KIF23 expression between ccRCC cell lines and normal kidney cell line.
This study aimed to explore the relationship between KIF23 and various cancer types, especially ccRCC, and constructed an effective prognosis prediction model for ccRCC patients, thus providing insight to a novel antitumor strategy.
Materials and Methods
Data Collection
Mutation, gene expression and clinical information data for 33 cancer types were downloaded from TCGA by using UCSC Xena13 (https://xena.ucsc.edu/). While Expression data for 31 tissues were obtained from GTEx (https://commonfund.nih.gov/GTEx). GSE2960914 gene expression data was retrieved from GEO database. We also collected 259 ferroptosis-related gene from FerrDb database.15 One thousand and six hundred forty-six KIF23-related genes were collected from Clinical Proteomic Tumor Analysis Consortium (CPTAC) database (https://cptac-data-portal.georgetown.edu/).
KIF23 Gene Expression Analysis
Differential expression of KIF23 between pan-cancer and normal tissues was analyzed by Oncomine database (http://www.oncomine.org)16 and the Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia2.cancer-pku.cn/#analysis). In the Oncomine database, the selection criteria were set to P<0.05 and fold-change of 1.5.
Analysis of the Relationships Between KIF23 and Prognosis, Clinical Phenotype, Immune Subtype
The correlation between overall survival and KIF23 expression was analyzed through univariate cox regression analysis. Forest plots were applied to show the P value, HR and 95% CI of each variable by the “forestplot” R package. Survival curves of different cancers were obtained using the Kaplan–Meier method. Tumor grade and stage were selected as significant clinical features. The relationship between KIF23 expression and stage, grade, immune subtype of pan-cancer was investigated by TISIDB database (http://cis.hku.hk/TISIDB/index.php). P<0.05 was considered as the criteria.
Analysis of KIF23 Genetic Alterations and Expression Profiling
cBioPortal database (http://www.cbioportal.org),17 which collected a multidimensional cancer genomics data set, was utilized to explore KIF23 genomic alterations including alternation frequency and type. Meanwhile, we evaluated overall survival (OS), disease-free survival (DFS) and progression free survival (PFS) of KIF23 gene alternation through Kaplan–Meier (KM) survival curves.
Analysis of the Relationships Between KIF23 and TMB, MSI, Immune Microenvironment and Immune Checkpoint Genes
TMB reflects somatic mutations in the exon region.18 The microsatellite instability (MSI) was calculated by R package of PreMSIm.19 The radar charts for TMB and MSI were finished by “Fmsb” R package. Estimation of Stromal and Immune Cells in Malignant Tumor Tissues Using Expression Data (ESTIMATE) was used to inferred stromal and immune microenvironment infiltration.20 The correlation of KIF23 expression with these scores in pan-cancer was displayed on the scatter plots. Meanwhile, we studied the correlation between KIF23 and immune checkpoint (ICP) genes in different cancers. And ICP genes were retrieved from previous study.21
Construction and Validation of Prognostic Model
KIF23-related genes (KRGs) were obtained from the UALCAN database (http://ualcan.path.uab.edu).22 Ferroptosis-Related Genes (FRGs) were collected from FerrDb database. Firstly, we retrieved the intersection between KIF23-related genes and FRGs. Second, the least absolute shrinkage and selection operator (LASSO) regression algorithm was applied to construct risk score model using R package “glmnet 4.0”. The KM survival analysis was employed to compare the overall survival between high-risk and low-risk groups. TimeROC analysis was applied to test the prediction accuracy of the model. Then, we used univariate COX analysis to determine independent prognostic factors for ccRCC OS. We regarded risk score as an independent predictor and built a nomogram associated with the risk score, age, stage and stage by R package “RMS” to predict the x-year overall recurrence rate.
In order to validate the nomogram, internal validation including C-index, calibration plot and decision curve analysis (DCA), were carried out based on TCGA data by using R package “RMS”. In addition, expression data of GSE29609 was used to perform external validation.
Gene Set Enrichment Analysis (GSEA)
To explore the functional role and potential signaling pathway of KIF23, GSEA was performed by GSEA v4.1.0. In the present study, gene size ≥100, FDR <0.15, nominal p< 0.05, |normalized enrichment score (NES)|> 1 and |enrichment score (ES)|> 0.60 were regarded as the selection criteria for meaningful pathways.
Quantitative Real-Time PCR
The expression pattern of KIF23 gene was evaluated in ccRCC cell lines including ACHN, 769-P and normal kidney cell line HK-2. These three cell lines were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). We extracted total RNA from cells using the HiPure Total RNA Mini Kit (Magen, China) and then quantified the RNA with NanoDrop. ReverTra Ace qPCR RT Kit (Toyobo, China) was used to reverse transcribed the RNA into cDNA. Finally, we carried out qRT-PCR with iQTM SYBR® Green Supermix (Bio-Rad). KIF23 forward primer: 5ʹ-AGTCAGCGAGAGCTAAGACAC-3ʹ, reverse primer: 5ʹ-GGTTGAGTCTGTAGCCCTCAG-3ʹ. GAPDH forward primer: 5ʹ-TGCACCACCAACTGCTTAG-3ʹ, reverse primer 5ʹ-GATGCAGGGATGATGTTC-3ʹ.
Results
Expression of KIF23 in Pan-Cancer
Firstly, we evaluated the expression of KIF23 in different tumors in comparison with normal tissues by Oncomine. The results indicated that KIF23 highly expressed in bladder cancer, brain and CNS cancer, breast cancer, cervical cancer, colorectal cancer, esophageal cancer, gastric cancer, head and neck cancer, liver cancer, lung cancer, lymphoma, melanoma, ovarian cancer, pancreatic cancer, prostate cancer and sarcoma (Figure 1A). Furthermore, in TCGA database and GTEx database, KIF23 expression was significantly elevated in various cancers, including adrenocortical carcinoma (ACC), bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), Ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell (TGCT), Thyroid carcinoma (THCA), uterine corpus endometrial carcinoma (UCEC), Uterine Carcinosarcoma (UCS) (Figure 1B).
Prognostic Value of KIF23 Across Pan-Cancer
In order to assess the potential prognostic value of KIF23, we employed univariate cox regression on the correlation between KIF23 expression and prognosis in pan-cancer. The results indicated that KIF23 was related to OS in ACC (P<0.0001), BLCA (p = 0.0167), KICH (p = 0.001), KIRC (P<0.0001), KIRP (P<0.0001), LGG (P<0.0001), LIHC (P<0.0001), LUAD (P = 0.0038), MESO (P<0.0001), PAAD (P<0.0001), PCPG (P = 0.004), PARD (P = 0.0174), THYM (P = 0.0098) (Figure 2A). Meanwhile, Kaplan–Meier survival analysis indicated that highly expressed KIF23 was associated with worse OS in ACC (P<0.0001, Figure 2B), BLCA (P = 0.0167, Figure 2C), KICH (P = 0.001, Figure 2D), KIRC (P<0.0001, Figure 2E), KIRP (P<0.0001, Figure 2F), LGG (P<0.0001, Figure 2G), LIHC (P<0.0001, Figure 2H), LUAD (P = 0.0038, Figure 2I), MESO (P<0.0001, Figure 2J), PAAD (P<0.0001, Figure 2K), PARD (P = 0.0174, Figure 2L), THYM (P = 0.0098, Figure 2M).
Correlation of KIF23 Expression with Clinicopathologic Features and Immune Subtypes
We studied the relevance of tumor grade and stage with KIF23 expression. The results suggested that KIF23 was highly expressed in high-grade groups compared with low-grade groups in KIRC (r = 0.285, P =2.78e-11, Figure 3A). KIF23 expression differed in tumor grade of various types of cancers. The details are shown in Supplementary Figure 1. The expression of KIF23 was associated with clinicopathologic stage in varieties of cancers (details in Supplementary Figure 2). For KIRC patients, high KIF23 expression was associated with high stage (r = 0.272, P =1.88e-10, Figure 3B). We also investigated the role of KIF23 on immune subtypes among pan-cancer by TISIDB website. There are six immune subtypes, including C1 (wound healing), C2 (IFN-gamma dominant), C3 (inflammatory), C4 (lymphocyte depleted), C5 (immunologically quiet) and C6 (TGF-b dominant). Consequently, KIF23 expression was correlated with different immune subtypes in various cancer types (Supplementary Figure 3). As for KIRC, KIF23 is significantly different in various subtypes (P =2.79e-07, Figure 3C).
 |
Figure 3 Association between KIF23 expression and tumor grade (A), stage (B) and immune subtypes (C) in KIRC. |
The Landscape of KIF23 Mutation Profile in Different Cancers
KIF23 genomic alterations in pan-cancer were analyzed across the cBioPortal website. We found that UCEC patients have the highest mutation frequency of KIF23, and the main alteration type was “mutation” (Figure 4A). Figure 4B shows the 3D structure of KIF23 protein. There were 131 mutation sites (including 112 missense, 12 truncating, 6 splice and 1 SV/fusion) between amino acids 0 and 960 (Figure 4C). The results also suggested that altered KIF23 group showed better prognosis in overall (P = 0.0449), disease-free (P = 6.244e-3) and progression-free (P = 0.0347) survival (Figure 4D).
Correlation of KIF23 Expression with TMB and MSI
TMB is closely associated with tumor prognosis and immunotherapy response. Our analysis indicated that KIF23 was significantly correlated with TMB in various cancer (Supplementary Figure 4A). In addition, KIF23 was related to MSI in different human cancer types (Supplementary Figure 4B).
Correlation of KIF23 Expression with Immune Microenvironment and Immune Checkpoints (ICP) Genes
We analyzed the correlation of KIF23 with stromal and immune score, which were obtained by The ESTIMATE algorithm, in 33 cancer types. The results suggested that KIF23 was significantly correlated with estimate score, stromal and immune score in various cancer types (Supplementary Figure 5A–C). As for KIRC, KIF23 was both positively associated with estimate score (R =0.28, P =5.87e-11 Figure 5A), immune (R =0.244, P =1.41e-08 Figure 5B) and stromal score (R =0.245, P =1.17e-08 Figure 5C). We also analyzed the correlation of KIF23 with ICP genes in pan-cancer. The result suggested that KIF23 expression was significantly associated with some ICP genes in several cancers (Supplementary Figure 5D).
 |
Figure 5 Correlation of KIF23 expression with estimate score (A), immune score (B) and stromal score (C) in KIRC. |
Construction of Risk Score Model for ccRCC
Firstly, we obtained 25 genes from the intersection between KIF23-related genes (KRGs) and FRGs (Figure 6). Then, we used LASSO regression algorithm for feature selection and 10-fold cross-validation to optimize the model (Figure 7A and B). Nine genes were screened out to construct a predictive risk score model, which was described as the formula: risk score = (0.3521*expression of BID) + (0.335*expression of KIF23) + (0.0135*expression of SLC1A5) + (−0.4725*expression of ATG7) + (0.1363*expression of CD44) + (0.2926*expression of ASNS) + (0.0232*expression of PSAT1) + (−0.306*expression of EIF2S1) + (0.1273*expression of SLC2A6). Risk scores were calculated, and the ccRCC samples was classified into high-risk and low-risk groups on the basis of the median of the risk scores (Figure 7C). Figure 7C also shows the survival status and time of the model group. Survival analysis indicated that the overall survival of patients in high-risk group is worse than that of low-risk group (P =4.87e-12, Figure 7D). Furthermore, we calculated the area under curve (AUC) of the time-dependent ROC curve to assess the prognostic value of the signature of these 9 genes. The AUC values of signature to predict the OS at 1-, 3-, and 5-year were 0.771, 0.73, and 0.74, respectively (Figure 7E), indicating great sensitivity and specificity of this model.
 |
Figure 6 Common genes between KIF23-related genes (KRGs) and ferroptosis-related genes (FRGs). |
Construction and Validation of Nomogram Model for ccRCC
Univariate COX analysis revealed that risk score, age, stage and grade could be the independent prognostic factors for ccRCC (Figure 8). Therefore, in order to build a clinically relevant method to predict prognosis in ccRCC patients, we developed a nomogram model to predict 1-, 3- and 5-year OS (Figure 9A). There were 4 predictive factors for the nomogram, including age, stage, grade and risk score. The C-index for the nomogram was 0.784. Decision-curve analysis (DCA) indicated that the net benefit of our model was greater than that of all screening or none-screening (Figure 9B). The calibration plots suggested that the predicted 1 -, 2 -, 3 -, and 5-year OS probabilities were consistent with the actual observations (Figure 9C). Subsequently, we employed GSE29609 data from GEO database as the external validation for our nomogram model. According to our nomogram model, the risk score and total points for samples in GSE29609 were calculated. GSE29609 samples were classified into high-risk and low-risk groups. Survival analysis suggested that patients in the high-risk group had worse OS (P = 0.0049, Figure 9D). The C-index for test set was 0.728. The calibration plots indicated that the predicted 1 -, 2 – and 3 -, OS probabilities were consistent with the actual observations in GSE29609, too (Figure 9E).
 |
Figure 8 Univariate Cox analysis for the independent prognostic factors for ccRCC. |
Gene Set Enrichment Analysis
To investigate the biological significance of KIF23 expression in ccRCC, we performed GSEA. The results revealed that KIF23 was significantly enriched in KEGG signaling pathways including cell cycle (nominal p < 0.001, n = 124, FDR < 15%, NES = 2.05, ES = 0.65, Figure 10A). And it was also enriched in GO signaling pathways including cell_cycle_G1_s_phase_transition (nominal p < 0.001, n = 108, FDR < 15%, NES = 2.07, ES = 0.67, Figure 10B), condensed chromosome (nominal p < 0.001, n = 181, FDR < 15%, NES = 2.07, ES = 0.65, Figure 10C), chromosome centromeric region (nominal p < 0.001, n = 163, NES = 2.01, FDR < 15%, ES = 0.67, Figure 10D), kinetochore (nominal p < 0.001, n = 109, FDR < 15%, NES = 196, ES = 0.68, Figure 10E).
 |
Figure 10 Gene set enrichment analysis. (A) KEGG pathway analysis of KIF23 in ccRCC. (B–E) GO functional annotation of KIF23 in ccRCC. |
Quantitative Real-Time PCR
The expression of KIF23 was further validated by qRT-PCR in ccRCC cell line. The results suggested that KIF23 mRNA levels were significantly higher in ccRCC cell line (ACHN and 769-P) than normal kidney cell line (HK-2) (Figure 11).
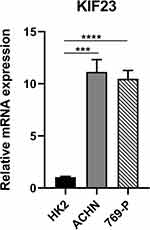 |
Figure 11 Validation of KIF23 mRNA expression in ccRCC and normal cell lines (***P < 0.0001, ****P < 0.00001). |
Discussion
ccRCC is a lethal urogenital tumor with high fatality rate and poor prognosis.23 There are many treatment options for ccRCC, including radical nephrectomy, renin sparing surgery, and immunotherapy, but the prognosis remains poor.24 Many molecularly targeted drugs have been used to treat patients with advanced tumors or cancer recurrence, which greatly improves survival time compared with chemoradiotherapy. However, the median DFS time and OS time of patients remain short due to the side effects of molecular targeted drugs and individual differences in patient sensitivity to drugs.25 Therefore, it is of vital importance to further explore more effective prognostic biomarkers and construct more accurate prognostic models.
In this study, we studied the correlation of KIF23 expression with prognosis, clinicopathologic features, immune subtype, TMB, MSI, immune infiltration and immune checkpoints (ICP) genes for pan-cancer. On the basis of the results of these analyses, we found that KIF23 was significantly associated with all aspects of ccRCC, implying that KIF23 could serve as a potential prognostic biomarker for ccRCC.
Ferroptosis is a ROS- and iron-dependent form of specialized programmed cell death, which occurs accumulation of lethal lipid peroxidation.26 Studies have indicated that ferroptosis can influence tumor progression and play a vital role in different cancers.27,28 Numerous genes have been shown to influence cancer formation and progression, particularly some of which contributed to aggressive malignancy by promoting ferroptosis.27,29
Therefore, we screened out FRGs associated with KIF23 and built a risk score model for ccRCC on the basis of the selected genes and KIF23. Meanwhile, we established a nomogram with age, stage, grade, and risk score as predictors to predict the survival of ccRCC patients. The external data GSE29609 further demonstrated that the prediction performance of our nomogram model was great. Finally, qRT-PCR also indicated the expression of KIF23 was higher in ccRCC cell lines compared with normal kidney cell line.
KIF23 was first found in 1992, which was a plus-end-directed motor enzyme.30 Studies have suggested KIF23 as a potential biomarker in several tumors.31 KIF23 promoted gastric carcinogenesis by activating Wnt/β-Catenin signaling.32,33 Knockdown of KIF23 inhibited breast cancer cell proliferation and induced apoptosis.34 Previous studies showed that DNA mutations in KIF23 can be found in half of cancer types with upregulated KIF23 expression and that CNAs of KIF23 are increased in 30% of tumors tested.35 Moreover, the p.p916r mutation KIF23 causes a rare inherited dyserythropoietic anemia (CDA III) with predisposition to blood cancer,36 and they further suggested that overexpression of KIF23 in non-small-cell lung cancer (NSCLC) may result from CNAs.37 The above studies show that KIF23 expression can be regulated by DNA mutations or CNAs. Therefore, KIF23 can be used as a new prognostic biomarker or cancer drug target for pan-cancer.
Several limitations of this research are as follows. First, we used an external dataset for validation, but this remains a retrospective study with inherent biases. Second, due to the limitation of public databases, the included clinical information was limited, and other variables that might affect the prognosis of patients were not included, such as specific treatment regimens. Therefore, more factors are needed to construct a more accurate nomogram. Furthermore, our study only used cell lines to verify KIF23 expression, and we still need further molecular experiments to verify and explore the function of KIF23.
Conclusions
In conclusion, by employing different bioinformatics methods, we found that KIF23 is closely associated with cancer initiation and prognosis. In ccRCC disease, KIF23 may be regarded as a potential prognostic biomarker, which contributes to the early diagnosis and treatment of the disease. Furthermore, a prognostic model based on KIF23 as well as KIF23-related FRGs is an important means to predict the OS of ccRCC patients. The nomogram of the present study may serve as a reliable tool to guide clinical work and better formulate personalized treatment regimens for ccRCC patients.
Data Sharing Statement
The dataset TCGA-KIRC for this study can be found in The Cancer Genome Atlas (http://cancergenome.nih.gov/). The datasets GSE29609 in this study can be found in the GEO (http://www.ncbi.nlm.nih.gov/geo/).
Ethical Statement
Our study was exempt from the approval of the Ethics Committee of Zhongnan Hospital of Wuhan University. The following are the related guidelines (Charter of the Medical Ethics Committee of Zhongnan Hospital of Wuhan University) which were followed:
“The responsibility of the ethics committee is to conduct independent, competent and timely review of the scientificity and ethical rationality of biomedical research projects involving people. The scope of the review includes:
- Using the methods of modern physics, chemistry, biology, traditional Chinese medicine and psychology to utilize human samples to study human physiology, psychological behavior, pathological phenomena, disease etiology and pathogenesis, as well as disease prevention, diagnosis, treatment and rehabilitation;
- Experimental research of new medical products on human body;
- The activities of collecting, recording, using, reporting or storing scientific research data such as human samples, medical records and behavior by means of epidemiology, sociology and psychology.”
The cell lines used in our research were purchased commercially and not obtained from human samples, which was not in the scope of the review. Therefore, our research was exempt from approval.
Acknowledgments
We would like to acknowledge the TCGA and GEO database developed by the National Institutes of Health (NIH).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the medical science and technology innovation platform support project of Zhong nan Hospital of Wuhan University (PTXM2019006).
Disclosure
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Vuong L, Kotecha RR, Voss MH, Hakimi AA. Tumor microenvironment dynamics in clear-cell renal cell carcinoma. Cancer Discov. 2019;9(10):1349–1357. doi:10.1158/2159-8290.CD-19-0499
3. Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387(10021):894–906. doi:10.1016/S0140-6736(15)00046-X
4. Cairns P, Srivastava S, Grizzle WE. Renal cell carcinoma. Cancer Biomark. 2010;9(1–6):461–473. doi:10.3233/CBM-2011-0176
5. Kotecha RR, Motzer RJ, Voss MH. Towards individualized therapy for metastatic renal cell carcinoma. Nat Rev Clin Oncol. 2019;16(10):621–633. doi:10.1038/s41571-019-0209-1
6. Barata PC, Rini BI. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67(6):507–524. doi:10.3322/caac.21411
7. Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359(6395):543–547. doi:10.1038/359543a0
8. Zhu C, Bossy-Wetzel E, Jiang W. Recruitment of MKLP1 to the spindle midzone/midbody by INCENP is essential for midbody formation and completion of cytokinesis in human cells. Biochem J. 2005;389(Pt 2):373–381. doi:10.1042/BJ20050097
9. Li XL, Ji YM, Song R, Li XN, Guo LS. KIF23 promotes gastric cancer by stimulating cell proliferation. Dis Markers. 2019;2019:9751923. doi:10.1155/2019/9751923
10. Takahashi S, Fusaki N, Ohta S, et al. Downregulation of KIF23 suppresses glioma proliferation. J Neurooncol. 2012;106(3):519–529. doi:10.1007/s11060-011-0706-2
11. Song X, Zhang T, Wang X, et al. Distinct diagnostic and prognostic values of kinesin family member genes expression in patients with breast cancer. Med Sci Monit. 2018;24:9442–9464. doi:10.12659/MSM.913401
12. Gao CT, Ren J, Yu J, Li SN, Guo XF, Zhou YZ. KIF23 enhances cell proliferation in pancreatic ductal adenocarcinoma and is a potent therapeutic target. Ann Transl Med. 2020;8(21):1394. doi:10.21037/atm-20-1970
13. Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678. doi:10.1038/s41587-020-0546-8
14. Edeline J, Mottier S, Vigneau C, et al. Description of 2 angiogenic phenotypes in clear cell renal cell carcinoma. Hum Pathol. 2012;43(11):1982–1990. doi:10.1016/j.humpath.2012.01.023
15. Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database (Oxford). 2020;2020. doi:10.1093/database/baaa021
16. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi:10.1593/neo.07112
17. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi:10.1158/2159-8290.CD-12-0095
18. Gonzalez-Pastor R, Goedegebuure PS, Curiel DT. Understanding and addressing barriers to successful adenovirus-based virotherapy for ovarian cancer. Cancer Gene Ther. 2021;28(5):375–389. doi:10.1038/s41417-020-00227-y
19. Li L, Feng Q, Wang X. PreMSIm: an R package for predicting microsatellite instability from the expression profiling of a gene panel in cancer. Comput Struct Biotechnol J. 2020;18:668–675. doi:10.1016/j.csbj.2020.03.007
20. Siemers NO, Holloway JL, Chang H, et al. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS One. 2017;12(7):e0179726.
21. Danilova L, Ho WJ, Zhu Q, et al. Programmed cell death ligand-1 (PD-L1) and CD8 expression profiling identify an immunologic subtype of pancreatic ductal adenocarcinomas with favorable survival. Cancer Immunol Res. 2019;7(6):886–895. doi:10.1158/2326-6066.CIR-18-0822
22. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
23. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi:10.1038/nrdp.2017.9
24. Rizzo A, Mollica V, Santoni M, et al. Impact of clinicopathological features on survival in patients treated with first-line immune checkpoint inhibitors plus tyrosine kinase inhibitors for renal cell carcinoma: a meta-analysis of randomized clinical trials. Eur Urol Focus. 2021. doi:10.1016/j.euf.2021.03.001
25. Vera-Badillo FE, Templeton AJ, Duran I, et al. Systemic therapy for non-clear cell renal cell carcinomas: a systematic review and meta-analysis. Eur Urol. 2015;67(4):740–749. doi:10.1016/j.eururo.2014.05.010
26. Xie Y, Hou W, Song X, et al. Ferroptosis: process and function. Cell Death Differ. 2016;23(3):369–379. doi:10.1038/cdd.2015.158
27. Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35(6):830–849. doi:10.1016/j.ccell.2019.04.002
28. Jiang L, Kon N, Li T, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520(7545):57–62. doi:10.1038/nature14344
29. Liang C, Zhang X, Yang M, Dong X. Recent progress in ferroptosis inducers for cancer therapy. Adv Mater. 2019;31(51):e1904197. doi:10.1002/adma.201904197
30. Zhao Z, Wang Z, Bao ZS, et al. Mutation and copy number alterations analysis of KIF23 in glioma. Front Genet. 2021;12:646929. doi:10.3389/fgene.2021.646929
31. Jian W, Deng XC, Munankarmy A, et al. KIF23 promotes triple negative breast cancer through activating epithelial-mesenchymal transition. Gland Surg. 2021;10(6):1941–1950. doi:10.21037/gs-21-19
32. Liang WT, Liu XF, Huang HB, Gao ZM, Li K. Prognostic significance of KIF23 expression in gastric cancer. World J Gastrointest Oncol. 2020;12(10):1104–1118. doi:10.4251/wjgo.v12.i10.1104
33. Liu Y, Chen H, Dong P, et al. KIF23 activated Wnt/β-catenin signaling pathway through direct interaction with Amer1 in gastric cancer. Aging (Albany NY). 2020;12(9):8372–8396. doi:10.18632/aging.103146
34. Zou JX, Duan Z, Wang J, et al. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res. 2014;12(4):539–549. doi:10.1158/1541-7786.MCR-13-0459
35. Sun X, Jin Z, Song X, et al. Evaluation of KIF23 variant 1 expression and relevance as a novel prognostic factor in patients with hepatocellular carcinoma. BMC Cancer. 2015;15:961. doi:10.1186/s12885-015-1987-1
36. Liljeholm M, Irvine AF, Vikberg AL, et al. Congenital dyserythropoietic anemia type III (CDA III) is caused by a mutation in kinesin family member, KIF23. Blood. 2013;121(23):4791–4799. doi:10.1182/blood-2012-10-461392
37. Vikberg AL, Vooder T, Lokk K, Annilo T, Golovleva I. Mutation analysis and copy number alterations of KIF23 in non-small-cell lung cancer exhibiting KIF23 over-expression. Onco Targets Ther. 2017;10:4969–4979. doi:10.2147/OTT.S138420
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





