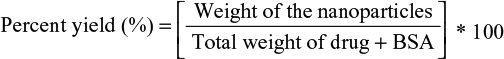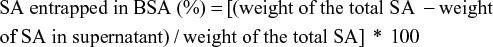Back to Journals » Nanotechnology, Science and Applications » Volume 10
Synthetic nanoparticles of bovine serum albumin with entrapped salicylic acid
Authors Bronze-Uhle ES, Costa BC, Ximenes VF, Lisboa-Filho PN
Received 12 July 2016
Accepted for publication 27 October 2016
Published 28 December 2016 Volume 2017:10 Pages 11—21
DOI https://doi.org/10.2147/NSA.S117018
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Israel (Rudi) Rubinstein
ES Bronze-Uhle,1 BC Costa,1 VF Ximenes,2 PN Lisboa-Filho1
1Department of Physics, São Paulo State University (Unesp), School of Sciences, Bauru, São Paulo, Brazil; 2Department of Chemistry, São Paulo State University (Unesp), School of Sciences, Bauru, São Paulo, Brazil
Abstract: Bovine serum albumin (BSA) is highly water soluble and binds drugs or inorganic substances noncovalently for their effective delivery to various affected areas of the body. Due to the well-defined structure of the protein, containing charged amino acids, albumin nanoparticles (NPs) may allow electrostatic adsorption of negatively or positively charged molecules, such that substantial amounts of drug can be incorporated within the particle, due to different albumin-binding sites. During the synthesis procedure, pH changes significantly. This variation modifies the net charge on the surface of the protein, varying the size and behavior of NPs as the drug delivery system. In this study, the synthesis of BSA NPs, by a desolvation process, was studied with salicylic acid (SA) as the active agent. SA and salicylates are components of various plants and have been used for medication with anti-inflammatory, antibacterial, and antifungal properties. However, when administered orally to adults (usual dose provided by the manufacturer), there is 50% decomposition of salicylates. Thus, there has been a search for some time to develop new systems to improve the bioavailability of SA and salicylates in the human body. Taking this into account, during synthesis, the pH was varied (5.4, 7.4, and 9) to evaluate its influence on the size and release of SA of the formed NPs. The samples were analyzed using field-emission scanning electron microscopy, transmission electron microscopy, Fourier transform infrared, zeta potential, and dynamic light scattering. Through fluorescence, it was possible to analyze the release of SA in vitro in phosphate-buffered saline solution. The results of chemical morphology characterization and in vitro release studies indicated the potential use of these NPs as drug carriers in biological systems requiring a fast release of SA.
Keywords: albumin nanoparticles, drug delivery, salicylic acid entrapped, nano-carriers
Introduction
Nanoparticle (NP) and microparticle carriers present an important drug delivery potential for the administration of therapeutic drugs. Systems with controlled release offer numerous advantages over conventional dosage forms, since they have better efficiency and low toxicity and provide convenience to the patient.1 The use of nanomaterials as pharmaceutical drug carriers to increase antitumor efficacy has been studied for more than 30 years.2 Initial clinical studies with liposomal nanocarriers were conducted in 1970.3 Currently, the use of nanoscale materials for drug delivery and diagnostics is in the forefront of medicine, since the encapsulation of a drug into NPs significantly improves its release profile in cells or tissues. By a proper synthesis, these nanomaterials can interact selectively with particular types of cells, passing through physiological barriers and penetrating deep into the tumor sites.4,5
In general, the use of nanocarriers can protect drugs from degradation and improve their function by facilitating the absorption and diffusion across the epithelium, modifying the distribution and pharmacokinetic profile of drugs in tissues and/or improving penetration and intracellular distribution.1,4–6 Several drug nanocarrier systems have been investigated in cancer treatment to minimize side effects and improve antitumor efficacy.
The performance of NPs depends on dimensionality, surface charge, surface modification, and hydrophobicity, which can affect the reactivity and physical and chemical properties of the material.1 Nanocarriers with optimized physical, chemical, and biological properties can penetrate the cells, where other larger materials would not be taken up or eliminated from the body. Smaller sizes (50–300 nm) enable interaction with biomolecules in the cell or on the cell surface, and thus nanocarriers can potentially affect cellular responses.7
Systems based on proteins have been studied for medicinal drug delivery, nutrient bioactive peptides, and probiotic organisms.7 Proteins are interesting materials since they possess the advantages of synthetic polymers, high adsorption capacity, low toxicity, biodegradability, and non-immunogenicity.
Human serum albumin (HSA) has a long history of pharmaceutical applications as a biodegradable carrier for the delivery of drugs. It is the most abundant protein in blood plasma, representing 52%–62% of total plasma proteins.8 It is a globular protein with dimensions of 4 × 4 × 14 nm spheroids, containing ~585 amino acids with a molecular weight of ~66,500 Da. The isoelectric point (IEP) of albumin is 4.5–5.0 and its surface is not charged. The main physiological function of albumin is to maintain the osmotic pressure and pH of the blood and still play an important role in the transport of a variety of endogenous and exogenous compounds, including fatty acids, metals, amino acids, steroids, and therapeutic drugs.9,10 Among other physiological functions, HSA controls the transport of drugs and nutrients through the human body and is also responsible for the effective deposition of drugs. It is easily available, stable, and of low cost. Historically, albumin has been extensively used as a biodegradable carrier of anticancer drugs due to its excellent biocompatibility and high stability in the blood and can accumulate in malignant or inflamed tissues.6
Bovine serum albumin (BSA) is highly water soluble and binds drugs and inorganic substances noncovalently. In addition, its structure is homologous to the three-dimensional structure of HSA. The main difference lies in the number of tryptophans (Trps). BSA has two Trps, while HSA has only one. This difference is useful in the context of its study by spectrofluorimetry, since this amino acid is the main one responsible for the intrinsic fluorescence of proteins.11–13
Microparticles and NPs of albumin can be obtained by the precipitation method in organic solvents, followed by a process of cross-linking with glutaraldehyde molecules (desolvation method).14
Transportation systems based on albumin NPs represent an important strategy since significant amounts of drug can be incorporated within the particle, depending on the drug-binding sites.12–17 Due to the well-defined structure of the protein containing charged amino acids, albumin NPs could favor the electrostatic adsorption of negatively or positively charged molecules, inside or on the surface, and the presence of hydrophobic cavities may facilitate the incorporation of water-insoluble drugs.18–20 BSA particles are small compared to microparticles and generally have more controlled properties for drug delivery when compared to liposomal nanocarriers.6,21,22
The first commercial product based on albumin NPs used in oncology was a 130 nm particle with bound paclitaxel.23,24 Currently, some studies have reported that particles of 20–200 nm are effective carriers for hydrophobic compounds.14
Salicylic acid (SA) and salicylates are components of various plants and have been used for medications with anti-inflammatory, antibacterial, and antifungal properties. Among its properties, SA has been considered a promising drug for the prevention of cardiovascular disease and cancer.25–27 SA and salicylates are the active metabolite of aspirin (acetyl SA) and have been used since the fifth century. However, these salicylic-derived drugs have a 50% bioavailability if orally administered, as they lose this activity following the initial step of deacetylation to SA.28–30
Recent reports in the specialized literature demonstrated that SA may be encapsulated or entrapped in different polymer matrices, and its release can be controlled by degradation and subsequent reabsorption of the polymer matrix of the sample by the human body.31–38 Thus, sustained and slow releasing of SA may be more appropriate for reducing the risk of cancer and cardiovascular diseases. The rapid release of SA may be desired for antimicrobial and anti-inflammatory applications, such as treatment of infections. Whereas release is totally dependent on the encapsulating matrix, the bioaccessibility of SA can be modified in the encapsulation process since it is related to and dependent on the chemical interaction between SA and the matrix.36,39
In this context, it is important to study and optimize these nanocarrier systems to better understand how to control drug release and its dependence on functional and biological aspects. As the interaction between the drug and albumin is limited by the surface aspects of these NPs, such as charge, binding sites, and amino acids, it is important to have a high ratio of drug to albumin, which may increase the drug concentration in target organs or tissues by decreasing the dose of drugs and toxic side effects.40 Furthermore, albumin has an unusual ability to bind various compounds, making it an excellent candidate for nanotechnology applications.
For the abovementioned reasons, the study of the synthesis and in vitro release of BSA NPs containing SA as the active agent was conducted. BSA NPs were synthesized by the desolvation method, where the size and surface charge of the NPs could be controlled by altering the pH in synthetic process. Therefore, the pH effect was evaluated on NP synthesis and the in vitro release of SA. The NPs were characterized by ultraviolet (UV)–visible, fluorescence and Fourier transform infrared (FTIR) spectroscopy. The analysis of zeta potential and dynamic light scattering (DLS) was used to investigate the physiochemical properties, namely size and charge for the SA–BSA NPs synthesized at different pHs. The structural surface morphology of BSA NPs and SA–BSA NPs was examined by field-emission scanning electron microscopy (FE-SEM) and transmission electron microscopy (TEM) showing a spherical smooth surface and SA entrapped in BSA NPs. The amount of unloaded SA and the release experiments were evaluated by fluorescence spectroscopy. The chemical and morphological characterization results and the in vitro release studies indicated the potential use of these NPs as drug carriers in biological systems requiring a fast release of SA.
Materials and methods
Reagent and chemicals
BSA (lyophilized powder, 66,000 kDa), 8% aqueous glutaraldehyde and SA were purchased from Sigma-Aldrich. Absolute ethanol was obtained from Synth. All chemicals were of analytical grade and were used without any further purification. All aqueous solutions were prepared in Milli-Q water. The pH of aqueous solutions was adjusted with 0.1 M HCl or 0.1 M NaOH using an Orion PerpHecT pH meter.
Preparation of BSA NPs and SA–BSA NPs
BSA NPs and SA–BSA NPs were prepared by the desolvation process.34,35
BSA NPs
BSA (200 mg) in 2 mL Milli-Q water (approximately pH 7.4) was stirred at 500 rpm at room temperature (25°C) for 10 minutes. The dissolved protein was transformed into NPs by the continuous addition of ethanol (1 mL/min), forming a turbid suspension. After 5 minutes, 0.16 mL of 8% (v/v) aqueous glutaraldehyde was added to cross-link the desolvated BSA NPs, and the reaction was kept under constant magnetic stirring for 18 hours. Next, the NP suspension was purified by three cycles of centrifugation at 12,000 rpm (Hermle Labortechnik GmbH) for 20 minutes to remove non-desolvated BSA, glutaraldehyde and ethanol. For each centrifugation step, NPs were redispersed in the same volume of deionized water (10 mL) by using an ultrasonication bath for 5 minutes. The obtained samples were then dried with nitrogen obtaining a nanosized powder.
SA–BSA NPs
In this procedure, 20 mg SA was dissolved in 2 mL Milli-Q water with the pH adjusted (5.4; 7.4; or 9), and 200 mg BSA were then added, followed by stirring for 10 minutes. Subsequently, the desolvating agent (absolute ethanol, 8 mL) was continuously added dropwise (1 mL/min) in solution with constant magnetic stirring at 500 rpm, resulting in the formation of a spontaneous opalescent suspension. After the addition of ethanol, 0.16 mL of 8% (v/v) aqueous glutaraldehyde was added to cross-link the desolvated BSA NPs, and the reaction was kept under constant magnetic stirring (500 rpm) for 18 hours at room temperature.
The resulting colloidal suspension was centrifuged at 12,000 rpm (Hermle Labortechnik GmbH) for 20 minutes. The supernatant was removed to obtain the particles. The samples obtained were purified by three cycles of centrifugation at 12,000 rpm for 20 minutes to remove non-desolvated BSA, free drug, glutaraldehyde, and ethanol.
For each centrifugation step, NP samples were redispersed in the small volume of deionized water (10 mL) by using an ultrasonication bath for 5 minutes. The samples were dried by solvent evaporation, resulting in an NP powder.
Synthesis yield and entrapment efficiency (EE%)
To determine the percent yield of synthesis, the NP powder sample obtained was weighed, and the percent yield was calculated using the following equation 1:
|
|
The amount of drug adsorbed or encapsulated in the albumin NPs was determined from the amount of free SA (weight [W]) in the supernatant after the centrifugation process, determined by fluorescence spectroscopy.
A standard calibration curve of peak maximum of fluorescence versus concentration, using known concentrations of SA in water, was plotted to determine free SA. The amount of SA loaded was determined using equation 2:
|
|
“In vitro” drug release experiments
To evaluate the release behavior of SA from the NPs, drug release experiments were performed in vitro. NPs (12 mg) were re-dispersed in 7 mL of phosphate-buffered saline (PBS, pH 7.4) incubated at 37±0.5°C under stirring at 100 rpm. At different times, 0.5 mL of the release medium was removed and another 0.5 mL of fresh PBS was supplied to maintain the total volume of the original solution at 7 mL. The aliquots removed were centrifuged, and the supernatant containing the drug (SA) was analyzed by fluorescence spectroscopy (excitation 330 nm), and all measurements were performed in triplicate. The cumulative drug release was plotted against time.
Characterization techniques
Physical characterization was performed using DLS and zeta potential, which measured size and surface charge of BSA NPs, respectively, using a Zetasizer Nano (Malvern, ZS 10, Malvern, UK) ZEN3601, with He–Ne laser, 633 nm. For measurements, BSA NP powder was suspended in absolute ethanol and Milli-Q water. Both dispersions were sonicated for 10 minutes to obtain uniformly dispersed NPs. The measurements were performed in triplicate at room temperature.
The structural morphology of BSA NPs was determined by FE-SEM (JSM-7500F; JEOL, Tokyo, Japan) and TEM (CM 200 Philips; Eindhoven, the Netherlands). For SEM measurements, NP solutions in ethanol (20 µL) were dripped and converted into powder on the surface of silicon substrates. Prior to analysis, samples were gold coated to make them electrically conductive and suitable for SEM.
For TEM analysis, 20–30 µL of the NP solutions in ethanol was dripped on a 200-mesh copper grid coated with carbon. The copper grid was allowed to dry for 2 hours at room temperature before observation.
UV–visible spectroscopy was used to analyze the NPs in aqueous solution (0.05 mg/mL), and the maximum absorption peak was measured by scanning 190–800 nm with a quartz cuvette of 1 cm path length, using a Shimadzu UV–visible spectrophotometer (UVmini-1240; Shimadzu, Kyoto, Japan).
Fluorescence spectroscopy was used to determine the presence and amount of free drug in solution to evaluate drug release. The fluorescence spectra were obtained using a microplate spectrofluorometer (Synergy H1 Hybrid reader; BioTek, Winooski, VT, USA) at room temperature, with excitation at 330 nm and emission in the range of 350–450 nm and with a fixed slit width of 5 nm. A standard calibration curve, fluorescence intensity at 405 nm versus concentration, was plotted to correlate fluorescence and concentration of free SA. The molecular structure and conformational changes in BSA due to interaction with SA were analyzed by FTIR spectroscopy. These measurements were performed using a Bruker Vertex 70 (Bruker; Ettlingen, Germany) in the region between 4000 and 400 cm-1, at room temperature in attenuated total reflectance (ATR) mode.19
Results and discussion
As discussed earlier, protein NPs have recently been the focus of research involving the interaction between albumin and other hydrophobic drug substances. The drug-binding properties of albumin NPs are clearly important for the understanding of reaction mechanisms, which provide a path for pharmacokinetic and pharmacodynamic mechanisms of these substances in various tissues. Studies conducted by Weber et al41 demonstrated that the process of protein NP formation depends on the amount of added desolvation agent, pH of the starting solution and amount of cross-linking agent (glutaraldehyde). The ethanol amount and pH of the solution directly alter NP size. Changing the amount of ethanol or rate of its addition in the process modifies the solubility of proteins, consequently altering the size of the NPs formed. Studies by Langer et al42 indicated that varying size of the NPs can be attributed mainly to the manual and dropwise addition of ethanol. Burns and Zydney43 investigated the effect of solution pH and showed that the loading surface has a strong effect on protein adsorption.
Changing pH significantly alters the net charge on the protein surface, since it consists of hydrogens of amino acids that interact with different ions in solution. The IEP of albumin is around 4.9, and at this pH the net charge on the protein surface is zero.44–46 At pH 4.9, there is a lack of electrostatic repulsion, and thus amorphous aggregates are readily formed through nonspecific interactions, mainly hydrophobic in nature.
At other pHs, positive charges may be generated by protonation of primary amino groups (particularly lysine), while negative charges are generated from deprotonation of carboxylic acid groups (glutamate and aspartate).44 Charges are generated on the whole surface of the protein, leading to greater electrostatic repulsion between molecules and decrease in hydrophobic interactions, thereby reducing aggregation and favoring a structural reorganization of the protein. Moreover, they can significantly alter the interaction and/or binding of compounds (drugs).
At pH above the IEP, negative charges and electrostatic forces dominate over hydrophobic interactions.45,\46 Another important component in the process of NP formation is the amount of cross-linking agent (glutaraldehyde) added. Glutaraldehyde does not change the size of the NPs but significantly alters the charge on the surface of the NPs obtained.42 Glutaraldehyde is a low-cost, water-soluble bifunctional reagent with high reactivity. The cross-linking of proteins or between protein molecules occurs by nucleophilic attack of the ε-amino groups of lysine and arginine residues in the protein by the two carbonyl groups of glutaraldehyde, forming Schiff bases in solution.47,48 The Schiff bases are unstable under acidic conditions but very stable at basic pH, aiding in the NP formation process. In addition, the lysine residues generally are not involved in the binding site, resulting in moderate preservation of the conformation of the protein and thereby the biological activity of the protein. As the amount of glutaraldehyde is linked to the number of free amino groups on the surface, it was kept constant during the study, since the amount of charge on the surface is associated with the efficacy of the interaction with the drug.48–51
For comparison purposes, the synthesis of drug-free NP albumin was also carried out at pH 7.4, according to the procedure described in the “Materials and methods” section. The protein NPs were obtained by the known process of desolvation. To do that, initially, BSA was placed in aqueous solution with the desired pH. In the case of the synthesis of NPs containing drugs, SA was also added to the albumin solution and allowed to interact using a magnetic stirring for 10 minutes. In this step, the charges formed on the surface of proteins by changing the pH could now interact differently with SA added. Ethanol (8 mL) was then added as desolvation agent at a controlled rate of 1 mL/min, which gradually decreases the solubility of proteins in solution leading to agglomeration. In this step, it was already possible to observe the formation of NPs, where some were partially soluble and stabilized in solution. To obtain NPs stabilized in solution with definite shape, glutaraldehyde was added, acting as a cross-linking agent. In this process, through covalent bonds, cross-linking agent was part of the protein surface NPs. The glutaraldehyde inserted into the protein, during the cross-linking process, has no toxicity, and the obtained NPs are non-toxic.45,47,48 However, the free compound in solution may exhibit toxicity. So, unreacted glutaraldehyde remaining in the solution was removed by centrifugation and purification of the NPs obtained.45,47,48,52
The purified NPs were dried by solvent evaporation under nitrogen flow. The yield of NPs and the entrapment efficiency (EE%) obtained were determined according to Equations 1 and 2, respectively, and are shown in Table 1.
  | Table 1 Percent yield and EE% of synthesis of BSA NPs and SA–BSA NPs Abbreviations: BSA, bovine serum albumin; EE, entrapment efficiency; NPs, nanoparticles; SA, salicylic acid. |
The low yield obtained for the reactions at pH 5.4 indicated that charges generated on the protein surface (close to BSA IEP) hampered drug entrapment into NPs.
Physical, chemical, and morphological characterization of NPs
After drying, pure BSA, BSA NPs, and SA–BSA NPs were analyzed by UV–visible absorption and fluorescence spectroscopy. These techniques provided physical characterization of the formation of the synthesized albumin NPs and presence of SA.
The UV–visible spectra of pure BSA, BSA NPs, and SA–BSA NPs were significantly different (Figure 1A) taking into account the position of maximum absorption peaks, when a spectral scan was evaluated between 190 and 800 nm. Figure 1B shows the fluorescence spectra, using an excitation wavelength of 280 nm and emission range of 310–450 nm during evaluation.53
Figure 1A shows that pure BSA had two absorption peaks at 192 and 278 nm. The intense peak at 192 nm is associated with the BSA backbone absorption, while the peak at 278 nm is associated with weak absorption of the aromatic amino acids phenylalanine (Phe), tyrosine (Tyr), and tryptophan (Trp).53,54 SA exhibited a strong absorption bands at 205, 228, and 296 nm. With the formation of BSA NPs, there was a strong decrease in the intensity and a blue shift for the absorption peak of 192 nm, and a slight increase for the peak at 278 nm, revealing that the changes related to the formation of BSA NPs mainly occurred in the BSA backbone bonds. It is noteworthy that the decrease in intensity in the SA may be linked to the fact that for concentration pure SA (0.05 mg/mL) and even SA–BSA NPs at a concentration of 0.05 mg/mL do not have the same amount of SA available. These results indicated that, in the formation of BSA NPs, glutaraldehyde modified mainly the BSA backbone amino acids, maintaining the aromatic amino acids Phe, Tyr, and Trp for drug interaction.53,54
According to chemical analysis, in the process of drug interaction with the NPs, the following can be considered: 1) SA interaction with BSA initially occurs in water at pH adjusted and 2) in the next step, cross-linking with glutaraldehyde occurs.
It is known that the main drug-binding region of BSA is localized in hydrophobic cavities of the IIA and IIIA subdomains, which exhibit similar chemical properties and are called sites I and II. These sites are mainly composed of amino acids to which molecules may bind on hydrophobic or positively charged surfaces. Thus, these two subdomains can specifically interact with negatively charged molecules or delocalized negative charges, such as heterocyclic ligands or carboxylic acids.53,54
Initially, when SA was added to the reaction medium, there was interaction at sites I and II, mainly with Tyr, Trp, and Phe, which absorb in the region of 278 nm.
SA–BSA NPs exhibited significant spectroscopic changes. They showed absorption in the regions of 202, 230, and 295 nm, while absorption at 278 nm decreased substantially, indicating the binding of SA to BSA in the NPs obtained.
As shown in Figure 1B, the fluorescence data corroborated the UV–visible results. It is known that BSA shows an optimal excitation wavelength at 278 nm, with maximum of emission around 350 nm, attributed to the amino acid residues of Tyr, Trp, and Phe. In contrast, some studies indicate that the intrinsic fluorescence can be attributed to Trp in BSA chain when the excitation is between 285 and 290 nm.55 In the BSA chain, there are two different types of Trp (134 and 212), localized in sites I and II, which can interact or bind with ligands. Trp-134 is localized on the protein surface and Trp-212 in the hydrophobic cavity of subdomain II. Thus, for pure BSA, BSA NPs, and SA–BSA NPs,56 the fluorescence wavelength changes were indicative of conformational and chemical changes in the BSA structure.57,58
The intrinsic fluorescence of BSA and SA, when excited at 280 nm, was obtained at 335 and 405 nm, respectively. Thus, according to the fluorescence spectra, the formation of BSA NPs leads to a decrease in the fluorescence intensity at 335 nm with a slight associated blueshift, indicating a conformational modification related to changes in the protein surface chain due to interaction with glutaraldehyde. The addition of SA at the beginning of the synthesis process leads to a strong decrease in the fluorescence intensity around 334 nm and an increase at 408 nm, characteristic emission of SA.11,59–63
The BSA NPs and SA–BSA NPs obtained were analyzed by FTIR spectroscopy. This technique has been used to evaluate the chemical and conformational changes that occur when NPs are formed or when they interact with other compounds through the slight shift in characteristic bands in the spectral regions of amide I and amide II.39 Figure 2 shows the FTIR spectra of pure BSA, BSA NPs, and SA–BSA NPs synthesized at pH 7.4 and 9.0.
  | Figure 2 Pure BSA, BSA NPs, and SA–BSA NPs FTIR spectra. Abbreviations: BSA, bovine serum albumin; FTIR, Fourier transform infrared; NPs, nanoparticles; SA, salicylic acid. |
Figure 2 shows the FTIR spectrum analysis, where it was possible to observe the major bands of pure BSA at 3280 cm-1 (amide A, related to N–H stretching), 2970 cm-1 (amide B, N–H stretching of  free ion), 1643 cm-1 (amide I, C=O stretching), 1515 cm-1 (amide II, related to C–N stretching and N–H bending vibrations), 1392 cm-1 (CH2 bending groups) and ~1260 cm-1 (amide III, related to C–N stretching and N–H bending). The most intense bands are associated with the secondary structure and conformation of proteins. The spectra of BSA NPs and SA–BSA NPs exhibited these characteristic bands of the protein and SA structure shifted slightly as shown in Table 2.45,64,65
free ion), 1643 cm-1 (amide I, C=O stretching), 1515 cm-1 (amide II, related to C–N stretching and N–H bending vibrations), 1392 cm-1 (CH2 bending groups) and ~1260 cm-1 (amide III, related to C–N stretching and N–H bending). The most intense bands are associated with the secondary structure and conformation of proteins. The spectra of BSA NPs and SA–BSA NPs exhibited these characteristic bands of the protein and SA structure shifted slightly as shown in Table 2.45,64,65
A small shift, of the absorption bands, was observed when compared to pure BSA with BSA NPs and SA–BSA NPs. The related changes in the amide I, II, and III bands confirm the formation of NP albumin and SA-loaded NP. In addition, some bands showed intensity differences (Figure 2), since it was possible to note a strong decrease intensities in the amide B (59% decrease) and amide III (40% decrease) bands, indicating changes in the C–N and/or NH bonds, due to interactions of different groups on BSA.
As discussed earlier, the pH change and the glutaraldehyde addition modify the surface charge of NPs, changing the electrostatic potential and colloidal stability of the protein NPs in solution.18,56,65 Thus, the electrostatic potential and the particle size were evaluated by zeta potential and DLS measurements by dispersion of the NPs in water and absolute ethanol. SA–BSA NPs synthesized at pH 7.4 showed respective zeta potential and DLS values of -6.45±1.23 mV and 182.20±12.20 d⋅nm for NPs dispersed in water and -33.2±1.90 mV and 81.48±0.9 d⋅nm for NPs dispersed in ethanol. For the SA–BSA NPs synthesized at pH 9, these values were 9.25±1.63 mV and 125.25±1.75 d⋅nm for NPs in water, and 32.8±4.3 mV and 76.54±0.46 d⋅nm for NPs in ethanol.18,56
The type of solvent used for dispersion strongly changes the colloidal stability of NPs. The NP solution in absolute ethanol exhibited a greater zeta potential and lower DLS value than the aqueous NP solution. The water pH changes the NP charge surface leading to the agglomeration of BSA NPs. A higher zeta potential value causes the particles to become stable by preventing their aggregation.
Morphological analysis of BSA NPs and SA–BSA NPs was carried out with FE-SEM, and the images obtained are shown in Figure 3A–C. Both types of synthesized NPs had a spherical morphology and an average size on a micrometric scale for BSA NPs (600±60 nm) and nanometric average, 110±7 nm and 138±6 nm, for SA–BSA NPs synthesized at pH 7.4 and 9.0, respectively. Furthermore, BSA NPs showed a greater nonuniform distribution, widely differing in size when compared to SA–BSA NPs.
Drug encapsulation by the protein during the synthesis process was also investigated by TEM, and the result obtained is shown in Figure 4. SA–BSA NPs showed a spherical morphology, as demonstrated by FE-SEM. Furthermore, it was possible to note the presence of structures with irregular surfaces inside these NPs, believed to be the encapsulated SA.
“In vitro” release studies
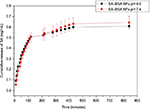
SA release from the protein NPs was analyzed by fluorescence measurement of suspensions of SA–BSA NPs in PBS, simulating a physiological environment at pH 7.4, and the results are shown in Figure 5.
The values of the release rate of NPs synthesized at pH 7.4 and 9.0 are provided in Table 3.
  | Table 3 Release rate of NPs (mg/mL⋅min) Abbreviations: BSA, bovine serum albumin; NPs, nanoparticles; SA, salicylic acid. |
A similar release behavior was seen for the samples obtained with the two synthesis procedures used, where there was an immediate release of SA, followed by a high release rate up to 120 minutes. Later, the release rate decreased and remained approximately constant from 400 minutes.
Conclusion
SA-loaded BSA NPs were synthesized with the desolvation process using glutaraldehyde cross-linking at different pHs. The pH change suggests that the process is associated with protein surface charges, generated at the beginning of the synthesis, and that this directly influences the entrapment process of SA, since synthesis was ineffective at pH 5.4. However, the pH slightly alters the release of SA from the protein NP. The release of SA occurs immediately, progressing to ~120 minutes. From this time, the release making constant is greatly reduced making constant starting 400 minutes, and these NPs may be applied in biological systems that require a rapid anti-inflammatory response.
Acknowledgments
The authors thank the Brazilian agencies CAPES, FAPESP, and CNPq for financial support, under contracts FAPESP 2014/204710 and CNPq 304810/2010-0. They are also grateful to Professor Marcelo Ornaghi Orlandi of the Department of Physical Chemistry, Instituto de Química, UNESP – Universidade Estadual Paulista, Araraquara, 14800-900, Brazil, for the FE-SEM and TEM analyses and to Dr Elaine Cristina Paris from EMBRAPA Instrumentação – Rua XV de Novembro, 1452, CP 741, CEP 13560-970, São Carlos, SP, Brazil, for zeta potential and DLS measurements. Dr A Leyva helped with English editing of the manuscript. Some parts of the results were presented as a poster in the 2015 MRS Fall Meeting and Exhibit, November 29, 2015, to December 4, 2015, Boston, MA, USA, and XIV Brazil MRS Meeting, September 27, 2015, to October 1, 2015, Rio de Janeiro, Brazil.
Disclosure
The authors report no conflicts of interest in this work.
References
Mehravar R, Jahanshahi M, Saghatoleslami N. Fabrication and evaluation of human serum albumin (HSA) nanoparticles for drug delivery application. Int J Nanosci. 2009;8(3):319–322. | ||
Brasseur F, Couvreur P, Kante B, et al. Actinomycin D absorbed on polymethylcyanoacrylate nanoparticles: increased efficiency against an experimental tumor. Eur J Cancer. 1980;16(11):1441–1445. | ||
Gregoriadis G, Neerunjun ED. Treatment of tumour bearing mice with liponsome-entrapped actinomycin D prolongs their survival. Res Commun Chem Pathol Pharmacol. 1975;10(2):351–362. | ||
Mohamed F, Van der Walle CF. Engineering biodegradable polyester particles with specific drug targeting and drug release properties. J Pharm Sci. 2008;97(1):71–87. | ||
Rasmussen JW, Martinez E, Louka P, Wingett DG. Zinc oxide nanoparticles for selective destruction of tumor cells and potential for drug delivery applications. Expert Opin Drug Deliv. 2010;7(9):1063–1077. | ||
Elzoghby AO, Samy WM, Elgindy NA. Albumin-based nanoparticles as potential controlled release drug delivery systems. J Control Release. 2012;157(2):168–182. | ||
Yana S, Zhanga H, Piao J, et al. Studies on the preparation, characterization and intracellular kinetics of JD27-loaded human serum albumin nanoparticles. Procedia Eng. 2015;102:590–601. | ||
Kopac T, Bozgeyik K, Yener J. Effect of pH and temperature on the adsorption of bovine serum albumin onto titanium oxide. Colloids Surf A. 2008;322:19–28. | ||
Brandes N, Welzel PB, Werner C, Kroh LW. Adsorption-induced conformational changes of proteins onto ceramic particles: differential scanning calorimetry and FTIR analysis. J Colloid Interface Sci. 2006;299(1):56–69. | ||
Huang BX, Kim HY, Dass C. Probing three-dimensional structure of bovine serum albumin by chemical cross-linking and mass spectrometry. J Am Soc Mass Spectrom. 2004;15(8):1237–1247. | ||
Tian J, Liu J, Tian X, Hu Z, Chen X. Study of the interaction of kaempferol with bovine serum albumin. J Mol Struct. 2004;691(1–3):197–202. | ||
Trnková L, Boušová I, Kubíček V, Dršata J. Binding of naturally occurring hydroxycinnamic acids to bovine serum albumin. Nat Sci. 2010;2(6):563–570. | ||
Xu H, Yao N, Xu H, Wang T, Li G, Li Z. Characterization of the interaction between eupatorin and bovine serum albumin by spectroscopic and molecular modeling methods. Int J Mol Sci. 2013;14(7):14185–14203. | ||
Xu R, Fisher M, Juliano RL. Targeted albumin-based nanoparticles for delivery of amphipathic drugs. Bioconjug Chem. 2011;22(5):870–878. | ||
Yang Z, Gong W, Wang Z, et al. A novel drug-polyethylene glycol liquid compound method to prepare 10-hydroxycamptothecin loaded human serum albumin nanoparticle. Int J Pharm. 2015;490(1–2):412–428. | ||
Patil GV. Biopolymer albumin for diagnosis and in drug delivery. Drug Dev Res. 2003;58(3):219–247. | ||
Irache JM, Merodio M, Arnedo A, Camapanero MA, Mirshahi M, Espuelas S. Albumin nanoparticles for the intravitreal delivery of anticytomegaloviral drugs. Mini Rev Med Chem. 2005;5(3):293–305. | ||
Mohanta V, Madras G, Patil S. Layer-by-layer assembled thin film of albumin nanoparticles for delivery of doxorubicin. J Phys Chem C. 2012;116(9):5333–5341. | ||
Wan X, Zheng X, Pang X, et al. The potential use of lapatinib-loaded human serum albumin nanoparticles in the treatment of triple-negative breast cancer. Int J Pharm. 2015;484(1–2):16–28. | ||
Subia B, Kundu SC. Drug loading and release on tumor cells using silk fibroin-albumin nanoparticles as carriers. Nanotechnology. 2013;24(3):035103. | ||
Tu H, Lu Y, Wu Y, et al. Fabrication of rectorite-contained nanoparticles for drug delivery with a green and one-step synthesis method. Int J Pharm. 2015;493(1–2):426–433. | ||
Wei Y, Li L, Xi Y, Qian S, Gao Y, Zhang J. Sustained release and enhanced bioavailability of injectable scutellarin-loaded bovine serum albumin nanoparticles. Int J Pharm. 2014;476(1–2):142–148. | ||
Nie S, Xing Y, Kim GJ, Simons JW. Nanotechnology applications in cancer. Annu Rev Biomed Eng. 2007;9:257–288. | ||
Kratz F. Albumin as a drug carrier: design of prodrugs, drug conjugates and nanoparticles. J Control Release. 2008;132(3):171–183. | ||
Gupta RA, Dubois RN. Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat Rev Cancer. 2001;1(1):11–21. | ||
Paterson JR, Lawrence JR. Salicylic acid: a link between aspirin, diet and the prevention of colorectal cancer. QJM. 2001;94(8):445–448. | ||
Amann R, Peskar BA. Anti-inflammatory effects of aspirin and sodium salicylate. Eur J Pharmacol. 2002;447(1):1–9. | ||
Erdmann L, Macedo B, Uhrich KE. Degradable poly(anhydride ester) implants: effects of localized salicylic acid release on bone. Biomaterials. 2000;21(24):2507–2512. | ||
Erdmann L, Uhrich KE. Synthesis and degradation characteristics of salicylic acid-derived poly(anhydride-esters). Biomaterials. 2000;21(19):1941–1946. | ||
Levy G. Comparative pharmacokinetics of aspirin and acetaminophen. Arch Intern Med. 1981;141(3):279–281. | ||
Whitaker-Brothers K, Uhrich K. Investigation into the erosion mechanism of salicylate-based poly(anhydride-esters). J Biomed Mater Res A. 2006;76(3):470–479. | ||
Tang SZ, June SM, Howell BA, Chai M. Synthesis of salicylate dendritic prodrugs. Tetrahedron Lett. 2006;47(44):7671–7675. | ||
Ji J, Hao S, Wu D, Huang R, Xu Y. Preparation, characterization and in vitro release of chitosan nanoparticles loaded with gentamicin and salicylic acid. Carbohydr Polym. 2011;85(4):803–808. | ||
Meng M, Feng Y, Guan W, Liu Y, Xi Y, Yan Y. Selective separation of salicylic acid from aqueous solutions using molecularly imprinted nano-polymer on wollastonite synthesized by oil-in-water microemulsion method. J Ind Eng Chem. 2014;20(6):3975–3983. | ||
Nowatzki PJ, Koepsel RR, Stoodley P, et al. Salicylic acid-releasing polyurethane acrylate polymers as anti-biofilm urological catheter coatings. Acta Biomater. 2012;8(5):1869–1880. | ||
Chandorkar Y, Bhagat RK, Madras G, Basu B. Cross-linked, biodegradable, cytocompatible salicylic acid based polyesters for localized, sustained delivery of salicylic acid: an in vitro study. Biomacromolecules. 2014;15(3):863–875. | ||
Dasgupta Q, Chatterjee K, Madras G. Combinatorial approach to develop tailored biodegradable poly(xylitol dicarboxylate) polyesters. Biomacromolecules. 2014;15(11):4302–4313. | ||
Dasgupta Q, Chatterjee K, Madras G. Controlled release of salicylic acid from biodegradable cross-linked polyesters. Mol Pharm. 2015;12(9):3479–3489. | ||
Rogers MA, Yan YF, Ben-Elazar K, et al. Salicylic acid (SA) bioaccessibility from SA-based poly (anhydride-ester). Biomacromolecules. 2014;15(9):3406–3411. | ||
Xie L, Tong W, Yu D, Xu J, Li J, Gao C. Bovine serum albumin nanoparticles modified with multilayers and aptamers for pH-responsive and targeted anti-cancer drug delivery. J Mater Chem. 2012;22(13):6053–6060. | ||
Weber C, Kreuter J, Langer K. Desolvation process and surface characteristics of HSA-nanoparticles. Int J Pharm. 2000;196(2):197–200. | ||
Langer K, Balthasar S, Vogel V, Dinauer N, Von Briesen H, Schubert D. Optimization of the preparation process for human serum albumin (HSA) nanoparticles. Int J Pharm. 2003;257(1–2):169–180. | ||
Burns DB, Zydney AL. Effect of solution pH on protein transport through ultrafiltration membranes. Biotechnol Bioeng. 1999;64(1):27–37. | ||
Eisele K, Gropeanu RA, Zehendner CM, et al. Fine-tuning DNA/albumin polyelectrolyte interactions to produce the efficient transfection agent cBSA-147. Biomaterials. 2010;31(33):8789–8801. | ||
Rohiwal SS, Satvekar RK, Tiwari AP, Raut AV, Kumbhar SG, Pawar SH. Investigating the influence of effective parameters on molecular characteristics of bovine serum albumin nanoparticles. Appl Surf Sci. 2015;334:157–164. | ||
Rohiwal SS, Pawar SH. Synthesis and characterization of bovine serum albumin nanoparticles as a drug delivery vehicle. Int J Pharm Bio Sci. 2014;5(4):51–57. | ||
Bhushan B, Dubey P, Kumar SU, Sachdev A, Matai I, Gopinath P. Bionanotherapeutics: niclosamide encapsulated albumin nanoparticles as a novel drug delivery system for cancer therapy. RSC Adv. 2015;5:12078–12086. | ||
Migneault I, Dartiguenave C, Bertrand MJ, Waldron KC. Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques. 2004;37(5):798–802. | ||
Okuda K, Urabe I, Yamada Y, Okada H. Reaction of glutaraldehyde with amino and thiol compounds. J Ferment Bioeng. 1991;71(2):100–105. | ||
Weetall HH. Immobilized enzymes: analytical applications. Anal Chem. 1974;46(7):602A–615A. | ||
Guisán JM. Aldehyde-agarose gels as activated supports for immobilization-stabilization of enzymes. Enzyme Microb Technol. 1988;10(6):375–382. | ||
Bhushan B, Gopinath P. Antioxidant nanozyme: a facile synthesis and evaluation of the reactive oxygen species scavenging potential of nanoceria encapsulated albumin nanoparticles. J Mater Chem B. 2015;3(24):4843–4852. | ||
Tian ZY, Song LN, Zhao Y, et al. Spectroscopic Study on the Interaction between Naphthalimide-Polyamine Conjugates and Bovine Serum Albumin (BSA). Molecules. 2015;20:16491–16523. | ||
Naveenraj S, Anandan S. Binding of serum albumins with bioactive substances – nanoparticles to drugs. J Photochem Photobiol C. 2013;14:53–71. | ||
Ni Y, Su S, Kokot S. Spectrofluorimetric studies on the binding of salicylic acid to bovine serum albumin using warfarin and ibuprofen as site markers with the aid of parallel factor analysis. Anal Chim Acta. 2006;580(2):206–215. | ||
Rohiwal SS, Tiwari AP, Verma G, Pawar SH. Preparation and evaluation of bovine serum albumin nanoparticles for ex vivo colloidal stability in biological media. Colloids Surf A Physicochem Eng Asp. 2015;480:28–37. | ||
Gentili PL, Ortica F, Favaro G. Static and dynamic interaction of a naturally occurring photochromic molecule with bovine serum albumin studied by UV−visible absorption and fluorescence spectroscopy. J Phys Chem B. 2008;112(51):16793–16801. | ||
Joshi P, Chakraborty S, Dey S, et al. Binding of chloroquine – conjugated gold nanoparticles with bovine serum albumin. J Colloid Interface Sci. 2011;355(2):402–409. | ||
Ju P, Fan H, Liu T, Cui L, Ai S. Probing the interaction of flower-like CdSe nanostructure particles targeted to bovine serum albumin using spectroscopic techniques. J Lumin. 2011;131(8):1724–1730. | ||
Liu XH, Xi PX, Chen FJ, Xu ZH, Zeng ZZ. Spectroscopic studies on binding of 1-phenyl-3-(coumarin-6-yl)sulfonylurea to bovine serum albumin. J Photochem Photobiol B. 2008;92(2):98–102. | ||
Wang YQ, Tang BP, Zhang HM, Zhou QH, Zhang GC. Studies on the interaction between imidacloprid and human serum albumin: spectroscopic approach. J Photochem Photobiol B. 2009;94(3):183–190. | ||
Hu YJ, Liu Y, Wang JB, Xiao XH, Qu SS. Study of the interaction between monoammonium glycyrrhizinate and bovine serum albumin. J Pharm Biomed Anal. 2004;36(4):915–919. | ||
Grdadolnik J. A FTIR investigation of protein conformation. Bull Chem Technol Maced. 2002;21:23–34. | ||
Kong J, Yu S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim Biophys Sin. 2007;39(8):549–559. | ||
Li C, Zhang D, Guo H, et al. Preparation and characterization of galactosylated bovine serum albumin nanoparticles for liver-targeted delivery of oridonin. Int J Pharm. 2013;48(1):79–86. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.