Back to Journals » International Journal of Nanomedicine » Volume 15
Synthesis of Silver Nanoparticles Using a Novel Cyanobacteria Desertifilum sp. extract: Their Antibacterial and Cytotoxicity Effects
Authors Hamida RS , Abdelmeguid NE, Ali MA, Bin-Meferij MM, Khalil MI
Received 14 November 2019
Accepted for publication 21 December 2019
Published 8 January 2020 Volume 2020:15 Pages 49—63
DOI https://doi.org/10.2147/IJN.S238575
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Supplementary video for ID 238575
Views: 1309
Reham Samir Hamida, 1 Nabila Elsayed Abdelmeguid, 1 Mohamed Abdelaal Ali, 2 Mashael Mohammed Bin-Meferij, 3 Mahmoud Ibrahim Khalil 4
1Molecular Biology Unit, Department of Zoology, Faculty of Science, Alexandria University, Alexandria, Egypt; 2Biotechnology Unit, Department of Plant Production, College of Food and Agriculture Science, King Saud University, Riyadh, Saudi Arabia; 3Department of Biology, College of Science Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia; 4Department of Biological Sciences, Faculty of Science, Beirut Arab University, Beirut, Lebanon
Correspondence: Mashael Mohammed Bin-Meferij; Mahmoud Ibrahim Khalil Tel +966 554477376; +961 81982014
Email [email protected]; [email protected]
Background: The emergence of multi drug-resistant (MDR) bacterial infections and cancer has necessitated the development and discovery of alternative eco-safe antibacterial and anticancer agents. Biogenic fabrication of metallic nanoparticles is an emerging discipline for production of nanoproducts that exert potent anticancer and antibacterial activity, and do not suffer from the limitations inherent in physiochemical synthesis methods.
Methodology: In this study, we isolated, purified, and characterized a novel cyanobacteria extract (Desertifilum IPPAS B-1220) to utilize in biofabrication of silver nanoparticles (D-SNPs). D-SNPs were produced by adding Desertifilum extract to silver nitrate solution under controlled conditions. Biofabrication of D-SNPs was confirmed using a UV-Vis spectrophotometer. The resultant D-SNPs were characterized using XRD, FTIR, SEM, and TEM. The toxicity of D-SNPs against five pathogenic bacteria and three cancer cell lines (MCF-7, HepG2, and Caco-2) was evaluated.
Results: Formation of D-SNPs was indicated by a color change from pale yellow to dark brown. The peak of the surface plasmon resonance of the D-SNPs was at 421 nm. The XRD detected the crystallinity of D-SNPs. FTIR showed that polysaccharides and proteins may have contributed to the biofabrication of D-SNPs. Under SEM and TEM, the D-SNPs were spherical with diameter ranges from 4.5 to 26 nm. The D-SNPs significantly suppressed the growth of five pathogenic bacteria, and exerted cytotoxic effects against MCF-7, HepG2, and Caco-2 cancer cells with IC 50 values of 58, 32, and 90 μg/mL, respectively.
Conclusion: These findings showed for the first time the potentiality of novel cyanobacteria strain Desertifilum IPPAS B-1220 to fabricate small SNPs that acted as potent anticancer and antibacterial material against different cancer cell lines and pathogenic bacterial strains. These findings encourage the researchers to focus on cyanobacteria in general and especially Desertifilum sp. IPPAS B-1220 for synthesizing different NPs that opening the window for new applications.
Keywords: nanotechnology, eco-friendly, D-SNPs, physicochemical, cytotoxic activity
Introduction
Nanoscience is developing rapidly, and many beneficial nanoproducts have been produced to benefit society.1 Metal nanoparticles (MNPs) exhibit unique optical and magnetic properties, have high surface area to volume ratios, and have low melting points and excellent mechanical strength, which allows for use of MNPs in medicine and industry.2 Moreover, these properties provide benefits in bio-imaging,3 diagnostics, and drug delivery.4 Among MNPs, silver nanoparticles (SNPs) have received the most attention due to their unique physicochemical and biological properties.5
Different physicochemical routes have been used to produce nanoparticles (NPs) with a variety of shapes and sizes, but the production of toxic by-products has limited use of these NPs.6 Advances in NP synthesis and increased use of these particles in many fields have led researchers to evaluate mechanisms to improve the biocompatibility of NPs.7,8 In addition, increased use of NPs has resulted in interest in the development of green approaches for the fabrication of NPs. Biological synthesis has several advantages including reduced production of toxic by-products, improved stability, and reduced toxicity against healthy cells.9,10 Furthermore, green synthesis is a low-cost approach, which allows for the rapid formation of efficient NPs.11 Also, comparing with the traditional methods, green methods enable the fabrication of different shapes of NPs that are distinguished with unique features under mild pressure and temperature conditions.9,11 Several green chemistry methods have been developed for the fabrication of NPs using plant extracts,12 algae,13 fungi,14 and bacteria.15 Cyanobacteria are excellent bio-systems for the intracellular or extracellular production of NPs. However, the biofabrication of metallic NPs using cyanobacteria has not been well characterized.16,17 Husain et al18 used the marine cyanobacterium Microchaete NCCU-342 to fabricate AgNO3 into smaller SNPs. Cepoi et al19 suggested that Spirulina Platensis and Nostoc linckia had potential for biofabrication of NPs because their cells contain numerous bioactive compounds. Desertifilum sp. was initially collected from biological crusts and identified as a filamentous cyanobacterial genus. Desertifilum sp. are oscillatorean cyanobacteria, which have filaments without differentiation of heterocytes and akinetes.20 These oscillatorean cells contain several bio-compounds such as pigments, proteins, and polysaccharides that can represent as potent reducing and capping materials during the formation of NPs.21
Overuse of antibiotics for the treatment of infectious diseases has contributed to the emergence of multidrug-resistant (MDR) bacteria.22 This issue has spurred the development of alternative neutral antibacterial agents with unique properties.23 Advances in nanotechnology have allowed for the production of MNPs, including SNPs, that exhibit low toxicity against healthy human cells and high bactericidal activity. Silver nanoparticles have potential as an alternative to antibacterial drugs due to promising effects against MDR bacteria in vivo and in vitro.24,25 Silver nanoparticles have been shown to exert a significant inhibitory activity toward gram-negative and gram-positive bacteria due to their small sizes and large surface area to volume ratios, which facilitate the extensive interactions between SNPs and bacteria.26
Currently, the scientific community is progressively developing to develop and discover novel therapeutic agents to treat cancer that is considered as the second leading cause of death worldwide.27,28 Cancer is a complex genetic disorder characterized by uncontrolled and abnormal cell growth.29 Chemotherapy and radiation are the primary treatments for tumors, but these therapies cause significant damage to normal cells.30 Advances in nanotechnology have allowed for development of MNPs that can be used for detection, prevention, and inhibition of cancer with low toxicity against normal cells.3 Aygun et al31 showed that after treatment MDA-MB-231 and HeLa cancer cells with Pt NPs, a significant cellular growth inhibition was observed. Bin-Meferij et al32 showed that SNPs synthesized using Nostoc sp. exhibited significant cytotoxicity against Caco-2 cells. Silver nanoparticles fabricated using Lyngbya majuscula showed antiproliferative activity against leukemia cell lines (K562, MOLT3, and REH).16 Recent publication showed that biogenic SNPs exhibited higher anticancer activity against MCF-7 and lower toxicity than 5-fluorouracil against normal cells.33
In this study, a novel cyanobacteria strain, Desertifilum IPPAS B-1220, was isolated, purified, cultured and genetically identified to use for green fabrication of silver NPs from silver nitrate. The resultant D-SNPs were physicochemical characterized and their bactericidal and antitumor activity was screened using five pathogenic bacterial strains and three cancer cell lines (MCF-7, HepG2, and Caco-2).
Materials and Methods
Materials
Silver nanoparticles were synthesized using Desertifilum sp. extracts. Silver nitrate (AgNO3) and cell culture materials were purchased from Sigma-Aldrich (St. Louis, MO, USA). Caco-2, MCF-7, and HepG2 were obtained from the Medical Research Institute (Alexandria, Egypt).
Cyanobacteria Isolation and Culture Conditions
Desertifilum sp. was previously isolated from a soil sample in Borg-El Arab city, Egypt, and purified by serial dilution as described by Bolch et al.34 Ten microliters of each diluted isolate was plated on BG11 nutrient agar plates and incubated at 25°C. The cultured strains were examined periodically using light and inverted microscopes to ensure that the cultured biomasses were pure.
The purified Desertifilum colonies were transferred to sterile 500 mL flasks containing BG-11 media (pH 7). The cultures were grown under a cool white fluorescent lamp that provided a light intensity of 2000 ± 200 Lux over a 12 h light/dark cycle at room temperature for 2 weeks.
Identification of Desertifilum IPPAS B-1220 sp
Morphological Examination
Light and inverted microscopes (Optika, Ponteranica BG, Italy) were used to identify the morphological features of the pure colonies, including shape, structure, and color. Taxonomic identification was performed based on morphology.35
Molecular Identification
Genomic DNA of the selected strain was extracted and purified as previously described by Singh et al.36 The DNA concentration was determined using a spectrophotometer (Jenway, Staffordshire, OSA, UK) at 260 nm. Agarose gel electrophoresis (0.8%; ReadyAgarose™ Precast Gel System Bio-Rad Laboratories, Inc., California, USA) was used to evaluate the integrity of the extracted genomic DNA.
Polymerase Chain Reaction (PCR) was used to determine the quality of the extracted genomic DNA template, and for 16s rDNA amplification using species-specific primers. Polymerase chain reaction was performed using a thermal cycler (Multigene Optimax, Labnet International, Inc., Edison, NJ, USA). The full length 1500 bp fragment of the 16S rRNA gene from the genome of the selected cyanobacterial isolate was amplified using PCR.37
The forward primer was 5ʹ-AGAGTTTGATCMTGGCTCAG-3ʹ (position 8 in the 16S rRNA gene according to E. coli numbering) and the reverse primer was 3ʹTACGGYACCTTGTTACGACTT-5ʹ (position 1514 in the 16S rRNA gene according to E. coli numbering). The thermal program was as follows: 4 min at 95°C followed by 30 cycles at 94°C for 1 min each (denaturation), then 55°C for 1 min (primer annealing), 72°C for 2 min (extension), and 10 min at 72°C. For standard PCR, the annealing temperature was set at 55°C. Following completion of the PCR reaction, a portion of the PCR product was quantified using agarose gel electrophoresis.38 The PCR products were purified to remove unincorporated nucleotides and excess primers using a PCR purification kit (Qiagen PCR Purification Kit, Hilden, Germany). The PCR product was mixed with 5 volumes of phosphate buffer (included in the kit), and the mixture was loaded onto a column. After centrifuging for 30 s at 13,000 rpm, the column was washed with 0.75 mL of TE buffer and centrifuged for 1 min to dry. The DNA was eluted using 50 µL of elution buffer. Gel electrophoresis was used to confirm successful DNA isolation, and the amplified DNA was stored in nuclease-free water and sequenced using an ABI 3730 DNA sequencer (Thermo Fisher Scientific, Waltham, MA, USA).
Sequence similarity was analyzed using a BLAST search (http://www.ncbi.nim.nih.gov/) after multiple alignments with sequences of closely related species. Comparative analysis was done utilizing CLUSTAL-W (https://www.genome.jp/tools-bin/clustalw) in the Bioedit program. BLAST and MEGA software were used to assess the similarity of the phylogenetic tree to other cyanobacteria species.
Preparation of Cyanobacteria Extracts
Desertifilum biomasses were harvested after 15 days by centrifugation at 4000 rpm for 20 min, and the pellets were washed at least twice with distilled water to remove any trace elements. The pellets were freeze-dried, and the dried pellets were crushed using a pestle and mortar to obtain a fine powder. An aqueous cyanobacteria extract was prepared by mixing 15 mg of fine powder with 15 mL of distilled water, then incubated at 40°C for 1 day. After 24 h, the aqueous extract was filtered using a Whatman filter No. 42 (Camlab, Cambridge, United Kingdom) and stored in a refrigerator at 4°C until use.
Production of SNPs by Cyanobacteria
Green synthesis of SNPs was performed by adding 90 mL of 1 mM aqueous AgNO3 to 10 mL of cyanobacteria filtrate at room temperature under direct illumination (2000 ± 200 Lux) for 24 h. A negative control was prepared by dissolving 90 mL of 1 mM aqueous AgNO3 in 10 mL of pure distilled water without cyanobacterial biomass. After 24 h, the mixture was centrifuged at 10,000 rpm for 15 min at 10°C, and the resultant pellets were washed with 10 mL of distilled water at least 3 times to remove any free biomass remnants or uncapped ligands. The pellets were spread on a sterilized petri dish and dried in an oven at 30°C for 1 day. The dried D-SNPs were scraped using a scalpel, and the fine powder was collected and stored in a sterile tube for analysis.
Characterization of D-SNPs
UV-Visible Spectroscopy
During the biological synthesis, and after 24 h, an aliquot of biofabricated D-SNPs was withdrawn and monitored using UV-VIS spectroscopy (UV1800 PC spectrophotometer, Shimadzu, Japan) across the wavelength range of 300 nm to 800 nm at a resolution of 1 nm.
X-Ray Diffraction
X-ray diffraction analysis (XRD) (X-ray 7000, Shimadzu, Japan) was used to evaluate the crystallinity of the biosynthesized SNP powder. The sample was drop-coated onto a silica plate by applying many layers of small amounts of D-SNPs on with intermittent drying. This resulted in a thick coat of SNPs, which was evaluated from 0° to 80° (2θ) using Cu Kα radiation generated at 30 kV and 30 mA with scan speed 4 deg/min.
Fourier-Transform Infrared Spectroscopy
The functional groups and composition of D-SNPs were determined using FTIR (Shimadzu, Japan). The D-SNP powders were diluted in potassium bromide (in the ratio 1:100) and the FT-IR instrument was operated in diffuse reflectance mode (DRS-800). Spectra were collected from 400 to 4000 cm−1 at a resolution of 4 cm−1. Functional groups were identified using reference spectra.
Scanning Electron Microscopy
Scanning electron microscopy (SEM) (Jeol, Tokyo, Japan) at 20 kV was used to characterize the shape and size of the resultant SNPs. The samples were coated with gold using a sputter coater (model: S150B, Edwards High Vacuum Ltd., England) to prevent the build-up of local electrical charges.
Transmission Electron Microscopy
Transmission electron microscopy (TEM, Jeol) was used to capture high-resolution, two-dimensional images of the biosynthesized SNPs. The morphology of the SNPs was visualized using TEM operated at an accelerating voltage of 200 kV. Samples were prepared by adding a drop of SNP suspension onto a carbon-coated copper grid and allowed to dry under an infrared lamp prior to analysis.
Cell Culture
Three different cancer cell lines were used; MCF-7 breast cancer cells, HepG2 liver cancer cells, and Caco-2 colorectal adenocarcinoma cells. Cells were maintained in Dulbecco’s Modified Eagle’s Minimum Essential Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 50 U/mL penicillin and streptomycin. The cells were incubated in a humidified atmosphere containing 5% CO2 at 37°C. Confluent cells were passaged using trypsin-EDTA.
D-SNP Cytotoxicity
The cytotoxicity of D-SNPs was screened against MCF-7, HepG2, and Caco-2 cells using the MTT assay as mentioned by Bin-Meferij et al.32 In brief, the malignant cells were cultured in 96-well plates at a density of 7 × 103 cells/well. After reaching confluency, the cells were subjected to increasing concentrations of 0.22-µm filtrated D-SNPs (31.25–1000 µg/mL). After 24 h, the DMEM medium was discarded and replaced with 100 µL of fresh culture medium, and 10 µL of 12 mM MTT stock solution was added to each well. Ten microliters of 12 mM MTT stock solution was added to 100 µL of medium as a negative control. The cells were incubated for 4 h at 37°C, then 100 µL/well DMSO was added, and the solutions were thoroughly mixed to dissolve the formazan crystals. Absorbance (A) was measured at 570 nm. The viability percentage was calculated using the formula:
(ATreated-ABlank)/(AControl-ABlank) × 100. The half-maximal growth inhibitory concentration (IC50) was determined using a sigmoidal curve.
Antibacterial Activity of D-SNPs
Microbial Strains and Culture
Five bacterial strains of gram-negative and gram-positive bacteria were obtained from King Khalid University hospital laboratory and the Department of Microbiology, King Saud University, Riyadh, Saudi Arabia. These strains included Bacillus cereus ATCC 10876, Pseudomonas aeruginosa ATCC 27853, Bacillus cercus ATCC 10876, Bacillus subtilis ATCC 6633, Shigella flexneri, and Salmonella enterica (clinical isolates). The five pathogens were grown in nutrient broth (Scharlab, S.L, SPAIN) at 37°C overnight and maintained through continuous sub-culturing in broth and on solid media.
Bactericidal Activity of D-SNPs Using Agar Well Diffusion Method
To evaluate the antibacterial activity of D-SNPs, 4 mL of bacterial saline mixture was mixed with 50 mL of warm nutrient agar (Merck, Darmstadt, Germany), and the mixture was poured into sterile plates and incubated for up to 18 h at 37°C. Four 8-mm wells were bored in each agar plate using a cork borer. One hundred microliters of D-SNPs suspension (1.54 mg/mL) was poured into three of the wells using a micropipette, and the blank solvent was used as a negative control. Other plates were treated with silver nitrate and streptomycin as a positive control. This experiment was repeated in a triplicate, and the mean and the standard error of the mean were calculated. The treated bacterial plates were incubated for 24 h at 37°C. The inhibitory activity of D-SNPs against bacteria was assessed by determining the diameter of the inhibition zone (IZ) in mm using a transparent ruler.
Statistical Analysis
All data are presented as a mean of three independent replicates (mean ± SEM), one-way analysis of variance (ANOVA) was performed for statistical analyses utilizing Prism 8.3 software (GraphPad Software Inc., La Jolla, CA, USA). Data were considered statistically significant at P < 0.01 and P < 0.001. For D-SNPs analysis, Origin8 (OriginLab Corporation, Northampton, MA, USA) and ImageJ (National Institutes of Health, Bethesda, MD, USA) were used.
Results and Discussion
Our hypothesis was “A novel cyanobacteria Desertifilum sp. will be biofabricate an ecofriendly silver nanoparticle that will have a biological activity against cancer cells and bacteria”
Desertifilum sp. showed a green or bright blue-green non-branching filamentous structure under a light microscope (Figure 1). Inverted light micrographs showed that the filaments were thin and long (more than 500 µm), as well as their widths ranged from 4 to 7 µm with an average width of 5.9 µm. Also, the filaments were present alone or organized in separate bundles or spiral trichomes of a dense mesh (Figure 2A). The filaments had apical ends as well as these filaments were motile non-constructed or slightly constricted at cross-walls (Video 1), the trichomes appear narrow at the end and were enclosed by a hyaline sheath firm (Figure 2B).39
 |
Figure 1 Low-power light micrograph of Desertifilum sp. Note: Filamentous appearance of Desertifilum sp. |
The resultant amplicon (1.5 kb) of the Desertifilum sp. is shown in Figure 3. The phylogenetic tree of Desertifilum sp. is shown in Figure 4. The AM2 strain was 99% similar to Desertifilum IPPAS B-1220.40 Desertifilum IPPAS B-1220 sequence was deposited in GenBank as MN629136.
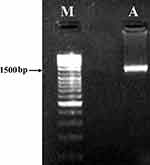 |
Figure 3 Electrophoresis of the resultant amplicon in 1% agarose gel. Notes: (A) 16S rDNA amplified using PCR from the genomic DNA of Desertifilum sp. (M) Molecular marker . |
 |
Figure 4 The phylogenetic tree of Desertifilum sp. |
To our knowledge, no previous studies have reported the use of Desertifilum sp. for the biosynthesis of nano-metals. Therefore, our study is the first to use this microorganism for the green synthesis of SNPs. The reduction process of AgNO3 to D-SNPs was characterized by a change in color from pale yellow to dark brown.41 The color change was due to the vibrations of surface plasmons in the resultant NPs.42 The D-SNPs had a peak at 421 nm (Figure 5), which indicated that D-SNPs were successfully synthesized from AgNO3. Sharpening of the SPR peak width is an indicator of nanometal size and monodispersity, and absorbance across the range of 320–580 nm is typical for SNPs.43,44 Ali et al45 found that the absorption peak of biogenic SNPs synthesized by Oscillatoria absorbed at 450 nm. Husain et al46 showed that SNPs synthesized from Oscillatoria sp. have an absorption peak at 485 nm. Furthermore, O-SNPs had a surface plasmon peak at 426 nm.47 Accordingly, the lower wavelength value indicated that smaller sized spherical NPs were formed,43 which suggested that Desertifilum sp. may produce particularly small nanoscale particles. Looking into the literature, our results indicated that Desertifilum sp. may be a promising candidate for biosynthesis SNPs.
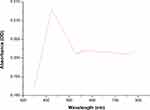 |
Figure 5 Ultraviolet-visible absorption spectra of D-SNPs. Note: Plasmon resonance of D-SNPs at 421 nm.Abbreviation: D-SNPs, silver nanoparticles synthesized by Desertifilum sp. |
The differences in SPR values may have been due to the different species of cyanobacteria used to produce SNPs,48 and the mechanisms of interaction between the cyanobacteria and NPs. Until now, the exact mechanism of green synthesis of NP is poorly understood, El-Batal et al49 reported that an NADH-dependent reductase was critical in SNP biofabrication. El-Naggar et al50 also revealed that cellular molecules such as proteins, enzymes, amino acids, carbohydrates, and photosynthetic pigments have been implicated in the fabrication process of silver ions.
Mahdieh et al51 suggested that the biofabrication of NPs is a two-step process. The first step involves adherence of SNPs to algal cells through electrostatic attraction between positively charged metal ions and negatively charged carboxylate ions on the algal surface. The second step involves the reduction of metal ions to MNPs by secreted reductase enzyme by the algal cells. The current data (Figure 5) may be interpreted based on that the cyanobacteria biomolecules such as proteins, enzymes, metabolites reduce silver nitrate into SNPs and/or phycobilisomes that mediated conformational changes to absorb light energy for initiating photosynthesis, this light absorption allows to transport the electrons from ground state to an excited state.52 So, under illumination may be silver nitrate incorporated into SNPs due to electrons moving between energy levels.
The range of 0° to 80° with 2ϴ values of 29.7,53,54 38.4,55 44.6,56 64.7,57 and 77.7,55 respectively, was associated with (310), (111), (200), (220), and (311) planes of silver (Figure 6). Each spectrum was compared with the available standard from the Joint Committee on Powder Diffraction Standards, and the biogenic D-SNPs were crystalline. The Scherrer equation, D = (kλ)/(β cos θ), was used to determine the crystallite size of the most intense peak at 2ϴ of 29.7. The crystallite size of the D-SNPs was 19.4 nm. Furthermore, interplanar spacing was 0.3 nm, as determined using the Bragg equation, d = λ/2 sin θ.
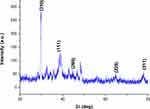 |
Figure 6 XRD pattern of D-SNPs. Abbreviations: XRD, x-ray diffraction; D-SNPs, silver nanoparticles synthesized by Desertifilum sp. |
Analysis using Fourier transform infrared (FTIR) showed that bioactive groups surrounded the biogenic silver nanoparticles. The FTIR spectrum of SNPs had six peaks at 601.81 cm−1, 1042.35 cm−1, 1626.05 cm−1, 2353.23 cm−1, and 3453.72 cm−1 for SNPs (Figure 7). The major peaks at 1042.35, 1626.05, and 3453.72 cm−1 indicated that the amides on the SNP surface might have included peptides and proteins present in the cell extract. Spectra of SNPs showed a strong broad absorption peak at 3453.72 cm−1, which is characteristic of O-H stretching vibration of the polysaccharide and N-H stretching vibration of proteins. A peak at 2353.23 cm−1 corresponded to CΞC stretching modes of vibration in alkyne groups. The absorption peak at 1626.05 cm−1 was assigned to bending N-H stretching vibration of amine groups of proteins and C=C stretching vibrations of alkenes. The band observed at 1042.35 cm−1 corresponded to the C-N stretching vibration of amines of proteins and C-O stretching vibration of vinyl ethers. The band at 601.81 cm−1 represented stretching of C-Br compounds. Based on the FTIR data, the presence of amide bonds of proteins and the hydroxyl group of polysaccharides indicated that the proteins and polysaccharides that surrounded silver may be responsible for the reduction of silver ion into SNPs and resulted in the formation of a capping layer around SNPs to prevent agglomeration and thereby the particles are stabilized. Our findings agreed with those of El-Naggar et al50 and Hamouda et al47 who revealed that biomolecules present in cell extracts could reduce Ag+ ions. Ahmad et al58 suggested that nitrate reductase may be a critical molecule in the reduction process of Ag+ ion to Ag0 by cyanobacteria grown on media containing NO3.
 |
Figure 7 FTIR spectra of green SNPs synthesized using Desertifilum sp. Abbreviations: FTIR, Fourier-transform infrared spectroscopy; SNPs, silver nanoparticles. |
Scanning electron micrographs showed that D-SNPs were roughly spherical and exhibited some agglomeration (Figure 8A). The size range of SNPs produced by Desertifilum sp. ranged from 22 to 40 nm with an average size of 31 nm (Figure 8B). Rasheed et al59 showed that SNPs fabricated using Convolvulus arvensis extract were spherical in shape and had an average size of 60 nm. These results indicated that Desertifilum sp. may be a promising microorganism for producing smaller SNPs.
Analysis using TEM showed that D-SNPs were spherical and were well dispersed without significant agglomeration. Particles ranged in size from 4.5 nm to 26 nm (Figure 9). The size range values agreed with the crystal sizes determined using XRD. Green SNPs were previously fabricated using Nostoc sp.,60 Synechococcus sp.,17 Anabaena sp.,61 Limnothrix sp., Synechocystis sp. 48–3, Anabaena doliolum sp.62 and Nostoc commune sp.63 with sizes ranging from 51 to 100, 15.2 to 266.7, 24.13, 31.86, 14.64, 10 to 50, and 15 to 54 nm, respectively. Yousefzadi et al64 reported that SNPs formed by the seaweed Enteromorpha flexuosa were 2–32 nm. The variations in sizes of SNPs produced by the same phylum may have been due to differences in silver reducing efficiency. Previous studies and our findings indicated that Desertifilum sp. has the potential to produce smaller SNPs.
This report demonstrated for the first time the anticancer activity of D-SNPs synthesized using cell extracts from Desertifilum sp. We evaluated the cytotoxic effects of D-SNPs against three human cell lines (MCF-7, HepG2, and Caco-2) using the MTT assay. The data revealed that D-SNPs significantly decreased the proliferation of the three different cancer cell lines in a dose-dependent manner compared with untreated cells (Figure 10A). The IC50 values of D-SNPs against HepG2, MCF-7, and Caco-2 cells were 32, 58, and 90 µg/mL, respectively (Figure 10B). The current data proved the efficiency of D-SNPs as an anticancer agent against different cell lines. The mechanisms by which SNPs inhibit cancer cell growth are poorly understood. However, a number of studies have shown that SNPs induced reactive oxygen species production, resulting in apoptosis.65,66 The current results showed that D-SNPs induced the greatest toxic effects against HepG2 cells. The higher sensitivity of HepG2 cells to D-SNPs may have been due to charge and the type of biomolecules surrounding D-SNPs. Baetke et al and El Nagar et al reported that SNP toxicity depended on size, concentration, and surface composition.4,33 Netala et al67 reported that surface charge and small size enhanced diffusion and dispersion into the tumor matrix. Bhatnagar et al68 showed that SNPs fabricated using extracellular pigment from Talaromyces purpurogenus have a significant cytotoxicity effect against liver cancer cell line. Abaza et al69 showed that Cs/PVA/Ag nanocomposites caused in killing 50% of HepG2 cells at 43.7 µg/mL. Based on these data, D-SNPs may be a potent antitumor agent against HepG2 with low IC50. Ranijitham et al70 reported that SNPs induced cytotoxicity in MCF-7 cells in a dose–response manner. Ramar et al71 showed that SNPs synthesized using fruits of Solanum trilobatum have an anticancer activity against MCF-7 cell line with IC50 equal to 300 µg/mL. This result indicates that D-SNPs may be a more efficient antitumor agent against MCF-7 cells. Martins et al72 showed that a polyelectrolyte complex containing SNPs exerted cytotoxic effects against Caco-2 cells, but not healthy African green monkey cells (VERO cells), at concentrations above 100 µg/mL. Comparing the chemically synthesized SNPs,69,72 D-SNPs showed a potent anticancer activity with low IC50 values and may be an alternative material to treat cancer.
This report evaluated for the first time the antibacterial activity of SNPs biosynthesized using Desertifilum sp. cellular extracts against different pathogenic bacteria strains. The antibacterial properties of D-SNPs against five bacterial strains (Bacillus cereus ATCC 10876, Pseudomonas aeruginosa ATCC 27853, Shigella flexneri, Salmonella enterica (clinical isolate), and Bacillus subtilis ATCC 6633) were evaluated using the agar well-diffusion assay. The data showed that D-SNPs induced varying degrees of toxicity against the tested bacteria (Figure 11). In addition, D-SNPs suppressed bacterial growth to a greater extent than silver nitrate, which was used as a positive control (Figure 12). As expected, the antibiotic streptomycin showed the highest antibacterial activity against five selected pathogens followed by D-SNPs (Table 1). Our results showed that D-SNPs inhibited the growth of Shigella flexneri to the greatest extent, as evidenced by an inhibition zone (IZ) diameter of 22.7 ± 0.3 while streptomycin resulted in IZ diameter equal to 26.3 ± 0.5. Also, for S.enterica the diameter of IZ measured after treating with D-SNPs was 16.7 ± 0.3 however the IZ diameter of streptomycin was 18.4 ± 0.4. These results are interesting since the values of IZ diameter caused by D-SNPs are nearly equals to the IZ value caused by streptomycin.
 |
Table 1 Agar Well-Diffusion Assay |
Although the D-SNPs have a potent inhibitory effect against B. cereus and B. subtilis with IZ diameter values of 17.3 ± 0.3 and 16.3 ± 0.3, respectively, the streptomycin was the most antibacterial agent that caused an IZ with a diameter of 29.5 ± 1.5 and 24.9 ± 0.5, respectively. D-SNPs inhibited the growth of Pseudomonas aeruginosa to a lesser degree, as evidenced by an inhibition zone diameter of 15 ± 0.0 (Table 1). In contrast, Pseudomonas aeruginosa were the most bacteria sensitive to AgNO3 treatment with IZ diameter of 14.7 ± 0.3, this value approximately the same as the IZ diameter resulted from treating the same bacteria with D-SNPs (Table 1). B. subtilis were the most bacteria that resisted the inhibitory action of silver nitrate with an IZ diameter of 9.5 ± 0.06.
These data indicate that D-SNPs have a significant antibacterial effect against gram-negative and gram-positive bacteria. Paul et al73 reported that SNPs synthesized extracellularly using Pseudomonas aeruginosa exerted cytotoxic activity against Shigella flexneri and Bacillus subtilis, as evidenced by inhibition zone diameters of 16.0 and 17.6 mm, respectively. El Nagar et al33 showed that the diameter of the inhibition zone of Pseudomonas aeruginosa treated with biogenic SNPs was 10 mm, and silver nitrate (5 mM) did not exert any antibacterial activity. Allafchian et al74 reported that SNPs synthesized using Phlomis leaf extract inhibited the growth of Bacillus cereus, with an inhibition zone diameter of 12.1 mm. In contrast, the treatment of Bacillus cereus with silver nitrate resulted in an inhibition zone diameter of 9.5 mm. Growth of Salmonella enterica was suppressed following treatment with SNPs fabricated using an aqueous extract of seaweed E. compressa, with an inhibition zone diameter of 10.5 mm (Table 2).75
 |
Table 2 The Antibacterial Activity of Different Biogenic SNPs Against Different Bacteria Strains |
The highest toxic effects of D-SNPs against Shigella flexneri may have been due to the thinner cell well of these gram-negative bacteria, allowing for better penetration of D-SNPs.76 Our results suggested that D-SNPs exhibited superior antibacterial effects due to the presence of surface biomolecules and small particle size.74 Romero et al77 explained that the biocompounds that surround the NP surface, charge and size of these particles play an important role in the attraction between bacterial cells and NPs and penetration of bacterial cell wall. Other publications are in accordance with our finding in which NPs with size less than 10 nm have the ability to penetrate the bacterial cell wall and interact with the bacterial biomolecule such as DNA, protein, lipids and sometimes with respiratory enzymes cause liberating of ROS such as hydrogen peroxide (H2O2), hydroxyl (OH–) and superoxide (O2− ) radicals that increase oxidative stress and damage nucleic acid and protein leading to death.78 Recent research by Sondi and Salopek-Sondi referred that the accumulation of SNPs (12 nm) on the cell wall of E. coli results in pit formation. Those pits cause a loss of outer membrane integrity, resulting in the release of cellular materials such as LPS molecules and proteins, and causing eventual cell death.79 The current findings indicated that D-SNPs may be potent antibacterial agents with a highly inhibitory effect against different pathogenic bacteria and may be used as alternative antibiotics to antibiotics that can be included in several products such as medical dressings and textile.
Conclusions
Cyanobacteria are an ancient group of photoautotrophic prokaryotes that contain valuable biomolecules such as pharmaceuticals, pigments, and proteins. Desertifilum sp. IPPAS B-1220 is a novel member of cyanobacteria that contain several biocompounds that can participate in SNP fabrication. In this study, we isolated, purified, cultured and molecular identified a novel cyanobacteria species (Desertifilum sp. IPPAS B-1220) and used their aqueous extract to synthesize silver nanoparticles. The biogenic SNPs detailed in this study were characterized using UV-Vis, FT-IR, XRD, SEM, and TEM. Physicochemical analyses showed that the D-SNPs were spherical and ranged in size from 4.5 to 26 nm. The presence of amides and hydroxyl groups indicated that proteins and polysaccharides were important factors in the biosynthesis of D-SNPs. The green D-SNPs produced in this study exerted potent antibacterial effects against gram-positive and gram-negative bacteria. Moreover, D-SNPs exerted significant cytotoxicity against MCF-7, HepG2, and Caco2. Our study showed that this approach was simple, green, efficient, and resulted in the production of SNPs at ambient temperature without the use of any toxic reducing or dispersing agents. Moreover, SNPs fabricated using Desertifilum sp. IPPAS B-1220 may be a promising antibacterial and anticancer agent that can be used in the medical and pharmaceutical industries. Further investigations are needed to demonstrate the optimal conditions to produce SNPs using Desertifilum sp. IPPAS B-1220. Additionally, future studies are needed to explain the biofabrication process using cyanobacteria and to explore the mechanism of action of biogenic SNPs against both bacteria and cancer cell lines.
Abbreviations
MNPs, metallic nanoparticles; NPs, nanoparticles; D-SNPs, silver nanoparticles synthesized by Desertifilum sp; SNPs, silver nanoparticles; AgNO3, silver nitrate; PCR, polymerase chain reaction; nm, nanometer; min, minute; µL, microliter; mg, milligram; mL, milliliter; µg, microgram; mM, millimolar; h, hour; cm−1, inverse centimeter; rpm, revolutions per minute; SEM, scanning electron microscope; TEM, transmission electron microscope; DLS, dynamic light scattering; FTIR, Fourier-transform infrared spectroscopy; XRD, X-ray diffraction; kV, kilovolt; kb, kilobase; SPR, surface plasmon resonance; ELISA, enzyme-linked immunosorbent assay; UV-vis, ultraviolet-visible spectroscopy; IZ, inhibition zone; platinum nanoparticles, Pt NPs.
Ethics Approval and Informed Consent
The current study was performed in accordance with the Research Ethical Committee guidelines published by the National Health and Medical Research Council and the Ministry of Health and Population in Egypt. The Department of Zoology (Science Faculty, Alexandria University) and the Department of Cancer Management and Research (Medical Research Institute, Alexandria University) granted permission for this work.
Data Sharing Statement
The data supporting this article are available in Figures 1–12. The data sets analyzed in the present study are available from the corresponding author upon reasonable request.
Acknowledgments
This research was funded by a grant for Scientists for Next Generation (ARST/SNG/FA/2014-5 no/5) from the Academy of Scientific Research and Technology, Ministry of Scientific Research, Egypt. This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-track Research Funding Program. The authors would like to thank Prof. Ramadan Awad for constructive comments of the manuscript.
Author Contributions
All authors contributed to data analysis, drafting, or revising the manuscript. All authors approved the final manuscript, and agree to be accountable for all aspects of the work.
Disclosure
This research was included in a request for patent 765805 from the Egyptian patent office, Academy of Scientific Research and Technology (ASRT), Egypt, submitted on 12/3/2019, titled “Biosynthesize of SNPs for the first time using new species of cyanobacteria Desertifilum IPPAS B-1220 and its tumor activity” http://www.egypo.gov.eg/. The authors report no other conflicts of interest in this work.
References
1. Sarker SD, Nahar L. Application of nanotechnology in phytochemical research. Pharm Sci. 2017;23(3):170–171. doi:10.15171/PS.2017.25
2. Horikoshi S, Serpone N. Microwaves in Nanoparticle Synthesis: Fundamentals and Applications. John Wiley & Sons; 2013.
3. Huerta-Aguilar CA, Ramírez-Guzmán B, Thangarasu P, Narayanan J, Singh N. Simultaneous recognition of cysteine and cytosine using thiophene-based organic nanoparticles decorated with Au NPs and bio-imaging of cells. Photochem Photobiol Sci. 2019;18:1761–1773. doi:10.1039/C9PP00060G
4. Baetke SC, Lammers T, Kiessling F. Applications of nanoparticles for diagnosis and therapy of cancer. Br J Radiol. 2015;88(1054):20150207. doi:10.1259/bjr.20150207
5. Chaloupka K, Malam Y, Seifalian AM. Nanosilver as a new generation of nanoproduct in biomedical applications. Trends Biotechnol. 2010;28(11):580–588. doi:10.1016/j.tibtech.2010.07.006
6. Gour A, Jain NK. Advances in green synthesis of nanoparticles. Artif Cells Nanomed Biotechnol. 2019;47(1):844–851. doi:10.1080/21691401.2019.1577878
7. Princy K, Gopinath A. Optimization of physicochemical parameters in the biofabrication of gold nanoparticles using marine macroalgae Padina tetrastromatica and its catalytic efficacy in the degradation of organic dyes. J Nanostruct Chem. 2018;8(3):333–342. doi:10.1007/s40097-018-0277-2
8. Tamarov K, Näkki S, Xu W, Lehto V-P. Approaches to improve the biocompatibility and systemic circulation of inorganic porous nanoparticles. J Mater Chem B. 2018;6(22):3632–3649. doi:10.1039/C8TB00462E
9. Thakkar KN, Mhatre SS, Parikh RY. Biological synthesis of metallic nanoparticles. Nanomedicine. 2010;6(2):257–262. doi:10.1016/j.nano.2009.07.002
10. Raja S, Ramesh V, Thivaharan V. Green biosynthesis of silver nanoparticles using Calliandra haematocephala leaf extract, their antibacterial activity and hydrogen peroxide sensing capability. Arab J Chem. 2017;10(2):253–261. doi:10.1016/j.arabjc.2015.06.023
11. Noruzi M, Zare D, Khoshnevisan K, Davoodi D. Rapid green synthesis of gold nanoparticles using Rosa hybridapetal extract at room temperature. Spectrochim Acta Part A. 2011;79(5):1461–1465. doi:10.1016/j.saa.2011.05.001
12. Aygün A, Gülbağça F, Nas MS, et al. Biological synthesis of silver nanoparticles using rheum ribes and evaluation of their anticarcinogenic and antimicrobial potential: a novel approach in phytonanotechnology. J Pharm Biomed Anal. 2019:113012–113038. doi:10.1016/j.jpba.2019.113012.
13. de Aragao AP, de Oliveira TM, Quelemes PV, et al. Green synthesis of silver nanoparticles using the seaweed Gracilaria birdiae and their antibacterial activity. Arab J Chem. 2016;2016:1–7. doi:10.1016/j.arabjc.2016.04.014
14. Aygün A, Özdemir S, Gülcan M, Cellat K, Şen F. Synthesis and characterization of Reishi mushroom-mediated green synthesis of silver nanoparticles for the biochemical applications. J Pharm Biomed Anal. 2019;112970–112991. doi:10.1016/j.jpba.2019.112970
15. Singh H, Du J, Singh P, Yi TH. Extracellular synthesis of silver nanoparticles by Pseudomonas sp. THG-LS1. 4 and their antimicrobial application. J Pharm Anal. 2018;8(4):258–264. doi:10.1016/j.jpha.2018.04.004
16. Roychoudhury P, Ghosh S, Pal R. Cyanobacteria mediated green synthesis of gold-silver nanoalloy. J Plant Biochem Biotechnol. 2016;25(1):73–78. doi:10.1007/s13562-015-0311-0
17. Keskin S, Oya N, Koçberber Kılıç N, Dönmez G, Tekinay T. Green synthesis of silver nanoparticles using cyanobacteria and evaluation of their photocatalytic and antimicrobial activity. J Nano Res. 2016.
18. Husain S, Afreen S, Yasin D, Afzal B, Fatma T. Cyanobacteria as a bioreactor for synthesis of silver nanoparticles-an effect of different reaction conditions on the size of nanoparticles and their dye decolorization ability. J Microbiol Methods. 2019;162:77–82. doi:10.1016/j.mimet.2019.05.011
19. Cepoi L, Rudi L, Chiriac T, et al. Biochemical changes in cyanobacteria during the synthesis of silver nanoparticles. Can J Microbiol. 2014;61(1):13–21. doi:10.1139/cjm-2014-0450
20. Dadheech PK, Abed RM, Mahmoud H, Mohan MK, Krienitz L. Polyphasic characterization of cyanobacteria isolated from desert crusts, and the description of Desertifilum tharense gen. et sp. nov. (Oscillatoriales). Phycologia. 2012;51(3):260–270. doi:10.2216/09-51.1
21. Shukla AK, Iravani S. Metallic nanoparticles: green synthesis and spectroscopic characterization. Environ Chem Lett. 2017;15(2):223–231. doi:10.1007/978-3-319-39303-2
22. Holmes AH, Moore LS, Sundsfjord A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. The Lancet. 2016;387(10014):176–187. doi:10.1016/S0140-6736(15)00473-0
23. Jain D, Daima HK, Kachhwaha S, Kothari S. Synthesis of plant-mediated silver nanoparticles using papaya fruit extract and evaluation of their anti microbial activities. Digest J Nanomater Biostruct. 2009;4(3):557–563.
24. Song JY, Kim BS. Rapid biological synthesis of silver nanoparticles using plant leaf extracts. Bioprocess Biosyst Eng. 2009;32(1):79–84. doi:10.1007/s00449-008-0224-6
25. Abdel-Raouf N, Al-Enazi NM, Ibraheem IBM, Alharbi RM, Alkhulaifi MM. Biosynthesis of silver nanoparticles by using of the marine brown alga Padina pavonia and their characterization. Saudi J Biol Sci. 2019;26(6):1207–1215. doi:10.1016/j.sjbs.2018.01.007
26. Abdel-Raouf N, Al-Enazi NM, Ibraheem IB. Green biosynthesis of gold nanoparticles using Galaxaura elongata and characterization of their antibacterial activity. Arab J Chem. 2017;10:S3029–S3039. doi:10.1016/j.arabjc.2013.11.044
27. Priyadarshini S, Gopinath V, Priyadharsshini NM, MubarakAli D, Velusamy P. Synthesis of anisotropic silver nanoparticles using novel strain, Bacillus flexus and its biomedical application. Colloids Surf B. 2013;102:232–237. doi:10.1016/j.colsurfb.2012.08.018
28. Varthamanan J. Anti cancer activity of silver nano particles bio-synthesized using stingless bee propolis (Tetragonula iridipennis) of Tamilnadu. Asian J Biomed Pharm Sci. 2015;5(40):30–38. doi:10.15272/ajbps.v4i40.654
29. Hassanpour SH, Dehghani M. Review of cancer from perspective of molecular. J Cancer Res Pract. 2017;4(4):127–129. doi:10.1016/j.jcrpr.2017.07.001
30. Brown K. Breast cancer chemoprevention: risk-benefit effects of the antioestrogen tamoxifen. Expert Opin Drug Saf. 2002;1(3):253–267. doi:10.1517/14740338.1.3.253
31. Aygun A, Gülbagca F, Ozer LY, et al. Biogenic platinum nanoparticles using black cumin seed and their potential usage as antimicrobial and anticancer agent. J Pharm Biomed Anal. 2019:112961–112982. doi:10.1016/j.jpba.2019.112961.
32. Bin-Meferij MM, Hamida RS. Biofabrication and antitumor activity of silver nanoparticles utilizing novel nostoc sp. Bahar M. Int J Nanomedicine. 2019;14:9019–9029. doi:10.2147/IJN.S230457
33. El-Naggar NE-A, Hussein MH, El-Sawah AA. Bio-fabrication of silver nanoparticles by phycocyanin, characterization, in vitro anticancer activity against breast cancer cell line and in vivo cytotoxicity. Sci Rep. 2017;7(1):10844–10864. doi:10.1038/s41598-017-11121-3
34. Bolch CJ, Orr PT, Jones GJ, Blackburn SI. Genetic, morphological, and toxicological variation among globally distributed strains of Nodularia (Cyanobacteria). J Phycol. 1999;35(2):339–355. doi:10.1046/j.1529-8817.1999.3520339.x
35. Anagnostidis K, Komárek J. Modern approach to the classification system of cyanophytes. 3-Oscillatoriales. Algolog Stud/Archiv Für Hydrobiologie. 1988; Supplement:327–472.
36. Singh SP, Rastogi RP, Häder D-P, Sinha RP. An improved method for genomic DNA extraction from cyanobacteria. World J Microbiol Biotechnol. 2011;27(5):1225–1230. doi:10.1007/s11274-010-0571-8
37. Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Paper presented at: Cold Spring Harbor symposia on quantitative biology. 1986.
38. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold spring harbor laboratory press; 1989.
39. Sinetova MA, Bolatkhan K, Sidorov RA, et al. Polyphasic characterization of the thermotolerant cyanobacterium Desertifilum sp. strain IPPAS B-1220. FEMS Microbiol Lett. 2017;364(4):fnx027–fnx037. doi:10.1093/femsle/fnx027
40. Nowruzi B, Khavari-Nejad R-A, Sivonen K, Kazemi B, Najafi F, Nejadsattari T. Identification and toxigenic potential of a Nostoc sp. Algae. 2012;27(4):303–313. doi:10.4490/algae.2012.27.4.303
41. Lengke MF, Fleet ME, Southam G. Biosynthesis of silver nanoparticles by filamentous cyanobacteria from a silver (I) nitrate complex. Langmuir. 2007;23(5):2694–2699. doi:10.1021/la0613124
42. Mulvaney P. Surface plasmon spectroscopy of nanosized metal particles. Langmuir. 1996;12(3):788–800. doi:10.1021/la9502711
43. Govindaraju K, Kiruthiga V, Kumar VG, Singaravelu G. Extracellular synthesis of silver nanoparticles by a marine alga, Sargassum wightii Grevilli and their antibacterial effects. J Nanosci Nanotechnol. 2009;9(9):5497–5501. doi:10.1166/jnn.2009.1199
44. Jena J, Pradhan N, Dash BP, Sukla LB, Panda PK. Biosynthesis and characterization of silver nanoparticles using microalga Chlorococcum humicola and its antibacterial activity. Int J Nanomater Biostruct. 2013;3(1):1–8.
45. Ali DM, Sasikala M, Gunasekaran M, Thajuddin N. Biosynthesis and characterization of silver nanoparticles using marine cyanobacterium, Oscillatoria willei NTDM01. Dig J Nanomater Biostruct. 2011;6(2):385–390.
46. Husain S, Sardar M, Fatma T. Screening of cyanobacterial extracts for synthesis of silver nanoparticles. World J Microbiol Biotechnol. 2015;31(8):1279–1283. doi:10.1007/s11274-015-1869-3
47. Hamouda RA, Hussein MH, Abo-elmagd RA, Bawazir SS. Synthesis and biological characterization of silver nanoparticles derived from the cyanobacterium Oscillatoria limnetica. Sci Rep. 2019;9(1):1–17. doi:10.1038/s41598-019-49444-y
48. Sathishkumar M, Sneha K, Yun Y-S. Immobilization of silver nanoparticles synthesized using Curcuma longa tuber powder and extract on cotton cloth for bactericidal activity. Biores Technol. 2010;101(20):7958–7965. doi:10.1016/j.biortech.2010.05.051
49. El-Batal A, Amin M, Shehata MM, Hallol MM. Synthesis of silver nanoparticles by Bacillus stearothermophilus using gamma radiation and their antimicrobial activity. World Appl Sci J. 2013;22(1):1–16. doi:10.5829/idosi.wasj.2013.22.01.2956
50. El-Naggar NE-A, Mohamedin A, Hamza SS, Sherief A-D. Extracellular biofabrication, characterization, and antimicrobial efficacy of silver nanoparticles loaded on cotton fabrics using newly isolated Streptomyces sp. SSHH-1E. J Nanomater. 2016;2016:1–17. doi:10.1155/2016/3257359
51. Mahdieh M, Zolanvari A, Azimee A. Green biosynthesis of silver nanoparticles by Spirulina platensis. Scientia Iranica. 2012;19(3):926–929. doi:10.1016/j.scient.2012.01.010
52. Fassioli F, Dinshaw R, Arpin PC, Scholes GD. Photosynthetic light harvesting: excitons and coherence. J R Soc Interface. 2014;11(92):20130901–20130923. doi:10.1098/rsif.2013.0901
53. Raza ZA, Rehman A, Anwar F, Usman A. Development and antibacterial performance of silver nanoparticles incorporated polydopamine–polyester-knitted fabric. Bull Mater Sci. 2016;39(2):391–396. doi:10.1007/s12034-016-1180-4
54. Granbohm H, Larismaa J, Ali S, Johansson L-S, Hannula S-P. Control of the size of silver nanoparticles and release of silver in heat treated SiO2-Ag composite powders. Materials. 2018;11(1):80–97. doi:10.3390/ma11010080
55. Labulo AH, Adesuji TE, Oseghale CO, et al. Biosynthesis of silver nanoparticles using Garcinia kola and its antimicrobial potential. Afr J Pure Appl Chem. 2016;10(1):1–7. doi:10.5897/AJPAC2015.0650
56. Sriramulu M, Sumathi S. Photocatalytic, antioxidant, antibacterial and anti-inflammatory activity of silver nanoparticles synthesised using forest and edible mushroom. Advan Nat Sci. 2017;8(4):045012–045022.
57. Shameli K, Ahmad MB, Zamanian A, et al. Green biosynthesis of silver nanoparticles using Curcuma longa tuber powder. Int J Nanomed. 2012;7:5603–5610. doi:10.2147/IJN.S36786
58. Ahmad A, Mukherjee P, Senapati S, et al. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium oxysporum. Colloids Surf B. 2003;28(4):313–318. doi:10.1016/S0927-7765(02)00174-1
59. Rasheed T, Bilal M, Li C, Nabeel F, Khalid M, Iqbal HM. Catalytic potential of bio-synthesized silver nanoparticles using Convolvulus arvensis extract for the degradation of environmental pollutants. J Photochem Photobiol B. 2018;181:44–52. doi:10.1016/j.jphotobiol.2018.02.024
60. Sonker AS, Pathak J, Kannaujiya VK, Sinha RP. Characterization and in vitro antitumor, antibacterial and antifungal activities of green synthesized silver nanoparticles using cell extract of Nostoc sp. strain HKAR-2. Can J Biotechnol. 2017;1(1):26–37. doi:10.24870/cjb.2017-000103
61. Patel V, Berthold D, Puranik P, Gantar M. Screening of cyanobacteria and microalgae for their ability to synthesize silver nanoparticles with antibacterial activity. Biotechnol Rep. 2015;5:112–119. doi:10.1016/j.btre.2014.12.001
62. Singh G, Babele PK, Shahi SK, Sinha RP, Tyagi MB, Kumar A. Green synthesis of silver nanoparticles using cell extracts of Anabaena doliolum and screening of its antibacterial and antitumor activity. J Microbiol Biotechnol. 2014;24(10):1354–1367. doi:10.4014/jmb.1405.05003
63. Morsy FM, Nafady NA, Abd-Alla MH, Elhady DA. Green synthesis of silver nanoparticles by water soluble fraction of the extracellular polysaccharides/matrix of the cyanobacterium Nostoc commune and its application as a potent fungal surface sterilizing agent of seed crops. Univ J Microbiol Res. 2014;2(2):36–43. doi:10.13189/ujmr.2014.020303
64. Yousefzadi M, Rahimi Z, Ghafori V. The green synthesis, characterization and antimicrobial activities of silver nanoparticles synthesized from green alga Enteromorpha flexuosa (wulfen) J. Agardh. Mater Lett. 2014;137:1–4. doi:10.1016/j.matlet.2014.08.110
65. Barcińska E, Wierzbicka J, Zauszkiewicz-Pawlak A, Jacewicz D, Dabrowska A, Inkielewicz-Stepniak I. Role of oxidative and nitro-oxidative damage in silver nanoparticles cytotoxic effect against human pancreatic ductal adenocarcinoma cells. Oxid Med Cell Longev. 2018;2018:1–15. doi:10.1155/2018/8251961
66. Gurunathan S, Raman J, Malek SNA, John PA, Vikineswary S. Green synthesis of silver nanoparticles using Ganoderma neo-japonicum Imazeki: a potential cytotoxic agent against breast cancer cells. Int J Nanomed. 2013;8:4399–4413. doi:10.2147/IJN.S51881
67. Netala VR, Bethu MS, Pushpalatha B, et al. Biogenesis of silver nanoparticles using endophytic fungus Pestalotiopsis microspora and evaluation of their antioxidant and anticancer activities. Int J Nanomed. 2016;11:5683. doi:10.2147/IJN.S112857
68. Bhatnagar S, Kobori T, Ganesh D, Ogawa K, Aoyagi H. Biosynthesis of silver nanoparticles mediated by extracellular pigment from talaromyces purpurogenus and their biomedical applications. Nanomaterials. 2019;9(7):1042–1062. doi:10.3390/nano9071042
69. Abaza A, Mahmoud G, Hegazy E, Amin M, Shoukry E, Elsheikh B. Cytotoxic effect of chitosan based nanocomposite synthesized by radiation: in vitro liver and breast cancer cell line. J Pharm Pharmacol. 2018;6:305–319. doi:10.17265/2328-2150/2018.04.002
70. Ranjitham AM, Suja R, Caroling G, Tiwari S. In vitro evaluation of antioxidant, antimicrobial, anticancer activities and characterisation of Brassica Oleracea. var. botrytis. L synthesized silver nanoparticles. Int J Pharm Pharm Sci. 2013;5(4):239–251.
71. Ramar M, Manikandan B, Marimuthu PN, et al. Synthesis of silver nanoparticles using Solanum trilobatum fruits extract and its antibacterial, cytotoxic activity against human breast cancer cell line MCF 7. Spectrochim Acta Part A. 2015;140:223–228. doi:10.1016/j.saa.2014.12.060
72. Martins AF, Follmann HD, Monteiro JP, et al. Polyelectrolyte complex containing silver nanoparticles with antitumor property on Caco-2 colon cancer cells. Int J Biol Macromol. 2015;79:748–755. doi:10.1016/j.ijbiomac.2015.05.036
73. Paul D, Sinha SN. Extracellular synthesis of silver nanoparticles using Pseudomonas aeruginosa KUPSB12 and its antibacterial activity. Jordan J Biol Sci. 2014;147(1573):1–6. doi:10.12816/0008246
74. Allafchian A, Mirahmadi-Zare S, Jalali S, Hashemi S, Vahabi M. Green synthesis of silver nanoparticles using phlomis leaf extract and investigation of their antibacterial activity. J Nanostruct Chem. 2016;6(2):129–135. doi:10.1007/s40097-016-0187-0
75. Ramkumar VS, Pugazhendhi A, Gopalakrishnan K, et al. Biofabrication and characterization of silver nanoparticles using aqueous extract of seaweed Enteromorpha compressa and its biomedical properties. Biotechnol Rep. 2017;14:1–7. doi:10.1016/j.btre.2017.02.001
76. Kim S-H, Lee H-S, Ryu D-S, Choi S-J, Lee D-S. Antibacterial activity of silver-nanoparticles against Staphylococcus aureus and Escherichia coli. Korean J Microbiol Biotechnol. 2011;39(1):77–85.
77. Romero-Urbina DG, Lara HH, Velázquez-Salazar JJ, et al. Ultrastructural changes in methicillin-resistant Staphylococcus aureus induced by positively charged silver nanoparticles. Beilstein J Nanotechnol. 2015;6(1):2396–2405. doi:10.3762/bjnano.6.246
78. Gurunathan S, Choi Y-J, Kim J-H. Antibacterial efficacy of silver nanoparticles on endometritis caused by Prevotella melaninogenica and Arcanobacterum pyogenes in dairy cattle. Int J Mol Sci. 2018;19(4):1210–1230. doi:10.3390/ijms19041210
79. Sondi I, Salopek-Sondi B. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. J Colloid Interface Sci. 2004;275(1):177–182. doi:10.1016/j.jcis.2004.02.012
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






