Back to Journals » Neuropsychiatric Disease and Treatment » Volume 18
Swimming Exercise Modulates Gut Microbiota in CUMS-Induced Depressed Mice
Authors Xie Y, Wu Z, Zhou L, Sun L, Xiao L, Wang G
Received 9 January 2022
Accepted for publication 21 March 2022
Published 5 April 2022 Volume 2022:18 Pages 749—760
DOI https://doi.org/10.2147/NDT.S355723
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Yuping Ning
Yumeng Xie,1 Zuotian Wu,1 Lin Zhou,1 Limin Sun,1 Ling Xiao,1,2 Gaohua Wang1,2
1Department of Psychiatry, Renmin Hospital of Wuhan University, Wuhan, Hubei, 430060, People’s Republic of China; 2Institute of Neuropsychiatry, Renmin Hospital of Wuhan University, Wuhan, Hubei, 430060, People’s Republic of China
Correspondence: Gaohua Wang, Department of Psychiatry, Renmin Hospital of Wuhan University, Jiefang Road No. 238, Wuhan, Hubei, 430060, People’s Republic of China, Tel +86 13607167402, Fax +86 027-88072022, Email [email protected]
Background: Gut microbiota is associated with anxiety and depression, while exercise has been proved to alleviate depressive symptoms. However, the interaction of exercise, depression, and gut microbiota remains unclear.
Methods: Male C57/BL6J mice were exposed to chronic unpredictable mild stress (CUMS) for 6 weeks and then were subjected to a 5-week swimming program. Behavioral tests, including sucrose preference test (SPT), open field test (OFT), elevated plus-maze (EPM) test, and tail suspension test (TST), were conducted to assess the anxiety-like and depressive behaviors. Gut microbiota analysis was carried out after sample collection.
Results: This study showed that CUMS induced depressive behaviors, but swimming exercise increased sucrose preference rate in the SPT, increased time in the center and number of rearing in the OFT, decreased time in the closed arm and increased time in the open arm in EPM, and decreased immobility time in the TST. Firmicutes were the predominant phylum in the gut microbiome, followed by the phyla Bacteroidetes and Proteobacteria. We further found that CUMS and swimming influenced the relative abundance of the genus Desulfovibrio, genus Streptococcus, genus p-75-a5. Among the metabolic pathways, aromatic biogenic amine degradation (PWY-7431), mono-trans and polycis decaprenyl phosphate biosynthesis (PWY-6383), chlorosalicylate degradation (PWY-6107), mycothiol biosynthesis (PWY1G-0), mycolyl-arabinogalactan-peptidoglycan complex biosynthesis (PWY-6397), toluene degradation I (aerobic) (via o-cresol) (PWY-5180), toluene degradation II (aerobic) (via 4-methylcatechol) (PWY-5182), and starch degradation III (PWY-6731) may be related to the mechanism of anti-depression effect.
Conclusion: Swimming exercise reverses CUMS-induced depressive behaviors, and the alteration of gut microbiota composition and regulation of microbiota metabolic pathways are involved.
Keywords: gut microbiota, swimming, chronic unpredicted mild stress, CUMS, depression, metabolic pathway
Introduction
Depression is a common mental illness characterized by low mood, anhedonia, and loss of interest. According to the Global Burden of Disease Project, depression is now the most serious disease burden among non-fatal neuropsychiatric disorders and is projected to be among top three disease burdens by 2030.1 According to the World Health Organization, approximately 350 million people suffer from depression, which can last for years.2
Previous studies have reported that depression, anxiety and exercise influence the composition of the gut microbiota. Research into the role of the gut microbiome in modulating brain function has rapidly increased over the past 10 years, implicating the microbiome as a possible key susceptibility factor for neurological disorders influencing emotional behavior and brain neurotransmitter systems.3–5 There is growing evidence linking the gut microbiome to depression and anxiety.6,7 Studies have shown that depression perturbs the gut microbiota at the abundance level, alters the fecal metabolic phenotype.6,8 Chronic unpredictable mild stress (CUMS) is a classic method for depression modeling in rodents. Studies9,10 have demonstrated that CUMS could not only lead to depression-like behavior but also change gut microbiota diversity and composition.
Evidence shows exercise has a regulatory effect on gut microbiota. Recent studies show that exercise can enhance the number of beneficial microbial species, enrich microbial-related metabolites, consequently improving the metabolic profile and immunological responses.11–13 However, there are few studies focusing on the role of gut microbiota in the antidepressant effect of exercise.
As the interaction of gut microbiota, depression and exercise is not fully revealed, we constructed mice depression models with/out exercise intervention to explore the alterations of composition and metabolic pathways of gut microbiota. Behavioral tests were utilized to evaluate the construction of our models and identify the anti-depression effect of exercise. We focused on the composition and related metabolism pathways of gut microbiota using 16S rRNA gene sequencing analysis and PICRUSt2 application. We are dedicated to identifying distinct species of gut microbiota that play a role in depressive-like behavior and exercise intervention models, and to proposing potentially meaningful metabolic pathways that might contribute to the pathogenesis of depression and the antidepressant mechanisms of exercise.
Materials and Methods
Animals
Five-week-old male C57/BL6J mice (n=24) were used in this study (provided by the animal facility at the Renmin Hospital of Wuhan University). Animals were fed under standard conditions (room temperature at 22 ± 1°C, 55 ± 5% humidity, and fed ad libitum) with a light cycle of 12 hours (lights on from 6:00 a.m. to 6:00 p.m.) and free access to food and water. The adaptive feeding period lasted for one week before the onset of the experiments. All procedures were conducted in accordance with the Regulations of Experimental Animal Administration issued by the State Committee of Science and Technology of the People’s Republic of China and the approval of the Ethics Committee of Renmin Hospital of Wuhan University. The animals were randomly divided into four groups: control and exercised mice (C+E, n=6), control and non-exercised mice (C+NE, n=6), CUMS and exercised mice (S+E, n=6), and CUMS and non-exercised mice (S+NE, n=6).
Chronic Unpredictable Mild Stress (CUMS)
A 6-week CUMS procedure was adopted. Briefly, a randomized schedule consisting of various stressors, including water deprivation (24 h), food deprivation (24 h), ice water swimming (5 min at 5°C), hot water swimming (5 min at 45°C), tail clamping (1 min), inversion of the light/dark cycle for 24 h, cage tilting (45°, 24 h) and damp bedding (24 h, 200 mL water per cage), was adopted. The mice were exposed to one of the stresses daily, and the same stress was not applied for two consecutive days to ensure the unpredictability of the experiment. Control mice were not exposed to any stress and were given free access to food and water.
Swimming Exercise Protocol
The swimming protocol consisted of two phases: adaptation and training. The training was graded beginning from 15 min on the first day until 60 min during the first week for adaptation. After adaptation, the protocol began with an intensity of 50 min/day, 5 d/week, for 4 weeks. The swimming program was performed in a large glass water tank (100 cm(L) × 60 cm(W) × 80 cm(H)) at 32 ± 1°C with a thermostat used to maintain the water temperature. The water depth was 60 cm so that the mice could not support themselves by touching the bottom with their feet; liquid soap was added to reduce surface tension and abolish floating behavior. The mice were swum in groups and continuously supervised. After swimming, the mice were toweled dry and kept warm. The protocol was carried out at the same time every day (between 9:00 and 11:00 a.m.).
Behavioral Tests
After the swimming exercise protocol, mice were subjected to several behavioral tests. The sucrose preference test (SPT), open field test (OFT), elevated plus-maze (EPM) test, and tail suspension test (TST) were conducted to assess the anxiety- and depression-like behaviors. All behavioral tests were performed from 8:00 a.m. to 12:00 p.m. in a quiet room, and the temperature and humidity of the testing room remained unchanged. For more details, please refer to Supplementary Methods.
Microbiome Sampling
Feces were collected directly from the cecum of mice into sterile tubes, which were immediately snap-frozen in liquid nitrogen and stored at −80°C.
DNA Extraction and PCR Amplification
Microbial community genomic DNA was extracted from all the samples using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to the manufacturer’s instructions. The DNA extract was analyzed on a 1% agarose gel, and DNA concentration and purity were determined using a NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA). The hypervariable region V3-V4 of the bacterial 16S rRNA gene were amplified with primer pairs 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) using an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, USA) (see Supplementary Methods).
Illumina MiSeq Sequencing
Purified amplicons were pooled in equimolar amounts and paired-end sequenced on an Illumina MiSeq PE300 platform/NovaSeq PE250 platform (Illumina, San Diego, USA) according to standard protocols by Wefind Biotechnology Co., Ltd. (Wuhan, China).
Processing of Sequencing Data and Statistical Analysis
The raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered using fastp version 0.20.0,14 and merged using FLASH version 1.2.715 with the following criteria: (i) the 300 bp reads were truncated at any site receiving an average quality score of <20 over a 50 bp sliding window, and the truncated reads shorter than 50 bp were discarded; reads containing ambiguous characters were also discarded; (ii) only overlapping sequences longer than 10 bp were assembled according to their overlapping sequence. The maximum mismatch ratio of the overlapping region is 0.2. Reads that could not be assembled were discarded. (iii) Samples were distinguished according to the barcode and primers, and the sequence direction was adjusted, exact barcode matching, and two nucleotide mismatches in primer matching.
Operational taxonomic units (OTUs) with a 97% similarity cut-off16 were clustered using UPARSE version 7.1,16 and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analyzed using the RDP Classifier version 2.217 against the 16S rRNA database (Silva v138) using a confidence threshold of 0.7.
Statistical analyses were performed using the SPSS software (ver. 26.0, SPSS Inc., Chicago, USA). The data were graphed using GraphPad Prism version 9.0, for MAC (San Diego, CA, USA). Statistical analysis of the quantitative multiple group comparisons was performed using Bonferroni test, two-way analysis of variance (ANOVA), or Kruskal–Wallis test. The confidence level was set at 95% (P < 0.05).
Results
Swimming Exercise Reversed Depression-Like Behavioral Changes Induced by CUMS
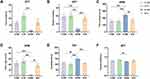
We examined the effect of swimming on depression-like behavioral changes induced by CUMS in mice. As shown in Figure 1A and B, the interaction effect was observed in number of rearing in the OFT (F (1,20) = 7.061, p = 0.015), CUMS reduced time in the center and number of rearing in the OFT (time in center: F (1,20) = 9.918, p = 0.005; number of rearing: F (1,20) = 15.888, p = 0.001), whereas swimming increased time in the center and number of rearing in the OFT (time in center: F (1,20) =12.623, p=0.003; number of rearing: F (1,20) = 9.467, p = 0.006); CUMS increased time in the closed arm in the EPM (F (1,20) = 15.782, p = 0.001), whereas swimming decreased it (F (1,20) =7.803, p=0.011, and the interaction effect was observed (F (1,20) = 5.971, p = 0.024) (Figure 1C). In contrast, CUMS decreased time in the open arm in the EPM (F (1,20) = 22.322, p < 0.001), whereas swimming increased it (F (1,20) = 10.631, p=0.004, and the interaction effect was observed (F (1,20) = 4.988, p = 0.037) (Figure 1D). In the TST, CUMS increased immobility time (F (1,20) = 10.730, p = 0.004), whereas swimming decreased it (F (1,20) = 16.207, p=0.001, and the interaction effect was observed (F (1,20) = 4.952, p = 0.038) (Figure 1E). In the SPT, CUMS decreased sucrose preference rate (F (1,20) = 5.851, p = 0.025), whereas swimming increased it (F (1,20) = 9.252, p=0.006) (Figure 1F), and no interaction effect was observed.
Post hoc Bonferroni test showed that CUMS induced depression-like behaviors in the S + NE group, including number of rearing in the OFT (p = 0.001, Figure 1B), increased time in the closed arm (p<0.001, Figure 1C) and decreased time in the open arm (p < 0.001, Figure 1D) in the EPM, increased immobility time in the TST (p=0.005, Figure 1E), compared with the C+NE group. Swimming exercise then reversed depression-like behaviors induced by CUMS in the S + E group, including increased number of rearing in the OFT (p=0.004, Figure 1B), decreased time in the closed arm, and increased time in the open arm in the EPM (p=0.008, Figure 1C; p=0.006, Figure 1D), decreased immobility time in the TST (p = 0.002, Figure 1E), compared to S+NE group.
CUMS and Swimming Exercise Modulated the Composition of the Gut Microbiome in Mice
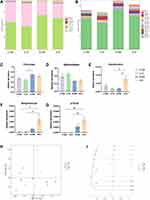
Changes in the gut microbiome were evaluated using 16S rRNA gene sequencing. First, the community diversity and richness were calculated using alpha diversity metrics. As shown in Figure 2, no significant differences were observed in community diversity estimators (Simpson index and Shannon index, Figure 2A and B) and richness estimators (Chao1 index and observed species index, Figure 2C and D) between the groups.
The number of OTUs detected at the domain, phylum, class, order, family, genus, and species levels was 173, 36, 21, 1238, 2949, 1816, and 355, respectively. At the phylum level, Firmicutes was the predominant phylum in the gut microbiome of mice, with a total abundance of nearly 60%, followed by the phyla Bacteroidetes, Proteobacteria and Actinobacteria (Figure 3A). Two-way ANOVA analysis showed that CUMS had a significant effect on the relative abundance of Firmicutes and Bacteroidetes (Firmicutes: F (1, 8) = 11.239 p = 0.010, Figure 3C; Bacteroidetes: F (1, 8) = 13.166 p = 0.007, Figure 3D), whereas swimming effected not significantly, and no interaction effect was observed. Post hoc Bonferroni test showed that no significant differences were observed in the relative abundance of Firmicutes and Bacteroidetes between the S + NE and the S + E groups.
At the genus level, Lactobacillus was the predominant genus in the gut microbiome of mice (Figure 3B). Three genera showed differences: Desulfovibrio, Streptococcus, and p-75-a5 (Figure 3E-G). Two-way ANOVA analysis showed that CUMS and swimming effected not significantly on the relative abundance of Desulfovibrio, but the interaction effect was observed (F (1, 8) = 12.965 p = 0.007); CUMS had a significant effect on the relative abundance of Streptococcus (F (1, 8) = 11.319 p = 0.010), whereas swimming effected not significantly, and no interaction effect was observed; CUMS had a significant effect on the relative abundance of p-75-a5 (F (1, 8) = 29.585 p = 0.001), whereas swimming effected not significantly, and no interaction effect was observed. Post hoc Bonferroni test showed the relative abundance of Desulfovibrio in the S+E group was significantly higher than that in the S+NE group (p = 0.027). In addition, compared with the C+E group, the relative abundance of Desulfovibrio, Streptococcus, and p-75-a5 was significantly higher than that in the S+E group (p = 0.022; p = 0.032; p = 0.005). We further analyzed the correlation between behavioral data and the composition of the gut microbiome, and visualized our results (Supplementary Figure S1).
A principal coordinate analysis (PCoA) plot based on unweighted UniFrac distances of gut microbiota from the four mice groups is shown (Figure 3H). Swimming exercise and CUMS induced a distinct structure of the gut microbiome, with 17.7% and 13.1% variation explained by the PC1 and PC2 principal components, respectively. In addition, hierarchical clustering analysis using the unweighted pair group method with arithmetic mean (UPGMA) demonstrated that most of the samples clustered in their groups (Figure 3I).
Swimming Exercise Modulated the Gut Metabolic Pathways in CUMS Mice
To further identify the relevant metabolic pathways involved in the swimming exercise and CUMS procedures, the differential metabolites were entered into the MetaCyc database to construct and analyze metabolic pathways. We calculated the abundance of the secondary functional pathways in the MetaCyc database (Figure 4A). Seven types of metabolic pathways- biosynthesis, degradation/utilization/assimilation, detoxification, generation of precursor metabolites and energy, glycan pathways, macromolecule modification, and metabolic clusters-were included. Among these pathways, the relative abundance of pathways involved in biosynthesis was the highest, followed by the generation of precursor metabolites and degradation/utilization/assimilation. Among the MetaCyc biosynthesis pathways, the relative abundance of nucleoside and nucleotide biosynthesis pathways was the highest, followed by the amino acid biosynthesis pathway. We then utilized MetagenomeSeq analysis to identify pathways with significant differences between the groups (Figure 4B-D). As shown in Figure 4B, the relative abundance of the five pathways- aromatic biogenic amine degradation (PWY-7431), mono-trans and polycis decaprenyl phosphate biosynthesis (PWY-6383), chlorosalicylate degradation (PWY-6107), mycothiol biosynthesis (PWY1G-0), and mycolyl-arabinogalactan-peptidoglycan complex biosynthesis (PWY-6397) in the S + E group were upregulated than those in the S + NE group (p < 0.01).
The relative abundance of secondary functional pathways in the KEGG database was also calculated (Supplementary Figure S2). Pathways were divided into six categories, including metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, and human diseases. The pathways are further classified into several levels. The results showed that the metabolism pathway had the highest relative abundance (Supplementary Figure S2A). Among these, carbohydrate metabolism was the highest, followed by amino acid metabolism. MetagenomeSeq analysis revealed significant differences between the groups (Supplementary Figure S2B-E). Styrene degradation pathways were significantly higher in the S+E group than those in the S+NE group (p < 0.001, Supplementary Figure S2B). Meanwhile, they were significantly lower in the S+NE group than those in the C+NE group (p < 0.001, Supplementary Figure S2C). In addition, the relative abundances of the linoleic acid metabolism, steroid biosynthesis and flavonoid biosynthesis pathways were different between the S+E and S+NE groups (p < 0.001, Supplementary Figure S2B).
Finally, the species composition of different pathways was analyzed based on pathways with significant differences in the MetaCyc database (Figure 5A-L). As shown in Figure 5, compared to the S+NE group, PWY-5180, PWY-5182, PWY-6107, PWY-6383, PWY-6397, PWY-6731, and PWY1G-0 were upregulated (Figure 5C-G, J, and L), and PWY-5028 was downregulated (Figure 5B) in the S+E group, suggesting that the species composition in these pathways may be related to the antidepressant mechanism of exercise. No discernible trend was observed in P221-PWY, PWY-6470, PWY-6562 and PWY-7431 (Figure 5A and H-K).
Discussion
Our results showed that CUMS-induced depression-like behaviors were reversed by a 4-week swimming exercise program. The alteration of gut microbiota composition and regulation of microbiota metabolic pathways are associated with depression-like behaviors and antidepressant mechanisms of exercise.
The CUMS protocol is a reliable and widely used depression model in rodents, and appears to be more effective than short-term stress exposure.18,19 The OFT evaluates an animal’s autonomous behavior, anxiety, and stereotypical behaviors such as rearing.20–22 Mice that prefer staying in the border can be described as showing signs of anxiety-like behavior, whereas mice with lower anxiety tend to spend more time in the center.23,24 The number of rearing indicated the exploratory activity of the mice. Generally, mice with fewer rearing condition are considered more anxious. The EPM test is used to test the anxiety-like behaviors and track the spontaneous behaviors of mice.25 A less anxious mouse will stay in the open arms of the maze longer, while a mouse with elevated anxiety tends to spend more time in the closed arms.26 The TST is a valid tool to reflect the depressive behaviors of mice,27 and the SPT is used to assess anhedonia in rodents. CUMS-induced depression mice display depression-like behaviors, including increased immobility time in the TST and decreased sucrose preference in the SPT.28 In our experiments, the results between the C+NE and S+NE groups suggested that CUMS induced depression-like and anxiety-like behaviors in mice.
The role of exercise as a non-pharmacological treatment of depression is gaining momentum, with findings showing that exercise alleviates depressive symptoms and also ameliorates body functions. There have been some animal studies that have demonstrated the effectiveness of exercise as a treatment for depression. For instance, Weina Liu et al29 demonstrated that four-week swimming exercise program alleviated the depressive behavior in male and female adult offspring. We chose a five-week swimming protocol, which was adapted from several previous studies.30,31 The results induced by the CUMS procedure were reversed by swimming exercise when comparing the S+E group with the S+NE group, which further explored the relation between the antidepressant benefits of exercise and gut microbiota.
To our knowledge, this is the first study to use 16S rRNA gene sequencing and metabolomic analysis to investigate the impact of swimming exercise on the gut microbiome and its metabolic profile in CUMS-stressed mice. Increasing evidence indicates that the gut community is associated with anxiety and depressive disorders,6,32 and growing attention has been paid to the role of microbiota in health and disease. It has been demonstrated that depression and anxiety are often accompanied by changes in colonic motility, which in turn alters the composition and stability of the gut microbiota.33 The regulatory mechanism of the gut microbiome on depression- and anxiety-like behaviors requires further study. We have previously shown that swimming exercise alleviates depression-like behavior in mice. We made a preliminary exploration, expecting to determine whether these outcomes are related to alterations in the composition and metabolites of gut microbiota.
First, we analyzed the community diversity and richness of the gut microbiome. No significant differences were observed in the community diversity and richness estimators between the groups. Previous studies34,35 have shown that mental induction of stress and depressive behaviors in rodents results in reduced gut microbiota richness and diversity. One clinical study36 found that no significant group differences in bacterial richness or diversity, and no significant group differences were observed at the phylum and genus levels. However, these inconsistent results require further investigation. Next, we investigated the composition of the gut microbiome. Generally, the bacterial phyla Firmicutes and Bacteroidetes represent > 90% of the intestinal community in healthy adults, and as such are correlated with the health outcomes in studies.37–39 In our results, Firmicutes was the predominant phylum in the gut microbiome of mice, with a total abundance of nearly 60%, followed by the phyla Bacteroidetes, Proteobacteria, Actinobacteria. At the genus level, Lactobacillus, a member of Firmicutes, was the predominant genus. Our results indicated that the alteration in the relative abundance of genus Desulfovibrio, genus Streptococcus, and genus p-75-a5, might be related to the mechanism of depression-like behaviors induced by CUMS. A recent study40 found that genus Desulfovibrio may play a potential role in the development of Parkinson’s Disease (PD), a progressive neurodegenerative disease, with genus Desulfovibrio present at higher levels in PD patients. However, there is no confirmed evidence proving the relation between the three genera, swimming and CUMS in the literature. The role of these four microorganisms in the antidepressant effect of swimming requires further experimental validation.
To identify the metabolic pathways in our study, we analyzed the abundance of microbial metabolic pathways in all samples using the MetaCyc database.41 Seven primary metabolic pathways were selected and analyzed. Among the biosynthesis pathways, the nucleoside and nucleotide biosynthesis pathways had the highest relative abundances. This pathway participates in the binding process of DNA or RNA, and the amino acid biosynthesis directly involved in protein synthesis also showed an extremely high abundance. Focusing on metabolite synthesis, we found that fatty acid and lipid biosynthesis, and carbohydrate biosynthesis were highly abundant. Fatty acid and lipid synthesis pathways were of great significance to depressed animals,42 and a long-term high-fat diet had been proven to be one of the means to induce a depression animal model.43
We then conducted a comprehensive analysis between different groups to screen out the significant up- or down-regulated pathways. Compared to the S+NE group, aromatic biogenic amine degradation (PWY-7431) in the S+E group showed the most significant upregulation. Studies44,45 have indicated that aromatic biogenic amine acids may have the antidepressant effects. A decrease in the degradation process may cause the accumulation of amines and decrease the levels of amino acids. Similarly, mono-trans and polycis decaprenyl phosphate biosynthesis (PWY-6383), chlorosalicylate degradation (PWY-6107), mycothiol biosynthesis (PWY1G-0), and mycolyl-arabinogalactan-peptidoglycan complex biosynthesis (PWY-6397) were downregulated. However, these metabolites lack valid confirmatory studies in animal models of depression. In addition, we can further explore the dominant flora involved in these pathways. For example, in PWY-5180 (Toluene Degradation I pathway), Corynebacterium made the largest contribution to it.
Depression and anxiety lead to neural circuit dysfunction in regions of the brain, such as the prefrontal cortex, thalamus, hippocampus.46–49 For example, previous results50,51 have shown that pulvinar is mutually connected with the sensory cortex, superior colliculus and amygdala and plays important roles in the multisensory integration and emotional response. And pulvinar dysfunction is associated with depression.49 It has been introduced above that the communication routes between the microbiota and the brain are related to various signaling and metabolites.52 With findings showing that gut microbes can modulate neurons through gut-brain circuits to promote metabolic benefits and alter body function and behaviors,53–56 we believe that alterations in the gut microbiota may also be linked to altered neural circuits in subjects with depression.
Our study has some limitations. Compared with the C+NE group, there was no significant difference in the abundance of Bacteroidetes and Firmicutes in the S+NE group. A previous study57 revealed a significant decrease in Bacteroidetes and a significant increase in Firmicutes in the CUMS group, and our study also proved that CUMS had significant effect on the two phyla. However, post hoc Bonferroni test showed no significance. The lack of statistical significance may be due to the small sample size and the difference in statistical method. We found that, using post hoc a least significant difference test, Bacteroidetes and Firmicutes indeed showed a consistent trend with Li’s work,57 under the impact of swimming. In addition, although we recognized the gut microbial communities and possible gut microbial metabolic pathways in CUMS mice, we did not examine additional depression model interventions such as social defeat stress model.58
Conclusion
This study showed that CUMS induced depressive behaviors, but swimming exercise increased sucrose preference rate in the SPT, increased time in the center and number of rearing in the OFT, decreased time in the closed arm and increased time in the open arm in EPM, and decreased immobility time in the TST. The CUMS-induced depression-like behaviors and anti-depression effect of swimming exercise may be related to the regulation of gut microbiota composition. We also identified several beneficial potential metabolic pathways. Our findings may help provide new clues for pathophysiological changes in depression and new insights into anti-depression therapy, especially the exercise intervention.
Abbreviations
CUMS, chronic unpredicted mild stress; OFT, open field test; EPM, elevated plus-maze; TST, tail suspension test; SPT, sucrose preference test; SEM, standard error of mean; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Funding
This study was supported by the National Natural Science Foundation of China (No.81871072 and NO. 82071523) and the Medical Science Advancement Program of Wuhan University (NO. TFLC2018001). Design of this study was supported by the Key research and development program of Hubei Province (2020BCA064).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442–e442. doi:10.1371/journal.pmed.0030442
2. Smith K. Mental health: a world of depression. Nature. 2014;515(7526):181. doi:10.1038/515180a
3. Cryan JF, O’Riordan KJ, Sandhu K, Peterson V, Dinan TG. The gut microbiome in neurological disorders. Lancet Neurol. 2020;19(2):179–194. doi:10.1016/S1474-4422(19)30356-4
4. Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest. 2015;125(3):926–938. doi:10.1172/JCI76304
5. Foster JA, McVey Neufeld KA. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36(5):305–312. doi:10.1016/j.tins.2013.01.005
6. Simpson CA, Diaz-Arteche C, Eliby D, Schwartz OS, Simmons JG, Cowan CSM. The gut microbiota in anxiety and depression - A systematic review. Clin Psychol Rev. 2021;83:101943. doi:10.1016/j.cpr.2020.101943
7. Mangiola F, Ianiro G, Franceschi F, Fagiuoli S, Gasbarrini G, Gasbarrini A. Gut microbiota in autism and mood disorders. World J Gastroenterol. 2016;22(1):361–368. doi:10.3748/wjg.v22.i1.361
8. Yu M, Jia H, Zhou C, et al. Variations in gut microbiota and fecal metabolic phenotype associated with depression by 16S rRNA gene sequencing and LC/MS-based metabolomics. J Pharm Biomed Anal. 2017;138:231–239. doi:10.1016/j.jpba.2017.02.008
9. Li H, Xiang Y, Zhu Z, et al. Rifaximin-mediated gut microbiota regulation modulates the function of microglia and protects against CUMS-induced depression-like behaviors in adolescent rat. J Neuroinflammation. 2021;18(1):254. doi:10.1186/s12974-021-02303-y
10. Wu J, Li J, Gaurav C, et al. CUMS and dexamethasone induce depression-like phenotypes in mice by differentially altering gut microbiota and triggering macroglia activation. Gen Psychiatr. 2021;34(6):e100529. doi:10.1136/gpsych-2021-100529
11. Donati Zeppa S, Agostini D, Gervasi M, et al. Mutual Interactions among Exercise, Sport Supplements and Microbiota. Nutrients. 2019;12(1):34. doi:10.3390/nu12010017
12. Mohr AE, Jäger R, Carpenter KC, et al. The athletic gut microbiota. J Int Soc Sports Nutr. 2020;17(1):24. doi:10.1186/s12970-020-00353-w
13. Monda V, Villano I, Messina A, et al. Exercise Modifies the Gut Microbiota with Positive Health Effects. Oxid Med Cell Longev. 2017;2017:3831972. doi:10.1155/2017/3831972
14. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi:10.1093/bioinformatics/bty560
15. Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi:10.1093/bioinformatics/btr507
16. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi:10.1038/nmeth.2604
17. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi:10.1128/AEM.00062-07
18. Antoniuk S, Bijata M, Ponimaskin E, Wlodarczyk J. Chronic unpredictable mild stress for modeling depression in rodents: meta-analysis of model reliability. Neurosci Biobehav Rev. 2019;99:101–116. doi:10.1016/j.neubiorev.2018.12.002
19. Qiao H, Li MX, Xu C, Chen HB, An SC, Ma XM. Dendritic Spines in Depression: what We Learned from Animal Models. Neural Plast. 2016;2016:8056370. doi:10.1155/2016/8056370
20. Katz RJ, Roth KA, Carroll BJ. Acute and chronic stress effects on open field activity in the rat: implications for a model of depression. Neurosci Biobehav Rev. 1981;5(2):247–251. doi:10.1016/0149-7634(81)90005-1
21. Prut L, Belzung C. The open field as a paradigm to measure the effects of drugs on anxiety-like behaviors: a review. Eur J Pharmacol. 2003;463(1–3):3–33. doi:10.1016/S0014-2999(03)01272-X
22. Kraeuter AK, Guest PC, Sarnyai Z. The Open Field Test for Measuring Locomotor Activity and Anxiety-Like Behavior. Methods Mol Biol. 2019;1916:99–103.
23. Crawley JN. Exploratory behavior models of anxiety in mice. Neurosci Biobehav Rev. 1985;9(1):37–44. doi:10.1016/0149-7634(85)90030-2
24. Bale TL, Contarino A, Smith GW, et al. Mice deficient for corticotropin-releasing hormone receptor-2 display anxiety-like behaviour and are hypersensitive to stress. Nat Genet. 2000;24(4):410–414. doi:10.1038/74263
25. Rodgers RJ, Dalvi A. Anxiety, Defence and the Elevated Plus-maze. Neurosci Biobehav Rev. 1997;21(6):801–810. doi:10.1016/S0149-7634(96)00058-9
26. Kraeuter AK, Guest PC, Sarnyai Z. The Elevated Plus Maze Test for Measuring Anxiety-Like Behavior in Rodents. Methods Mol Biol. 2019;1916:69–74.
27. Stukalin Y, Lan A, Einat H. Revisiting the validity of the mouse tail suspension test: systematic review and meta-analysis of the effects of prototypic antidepressants. Neurosci Biobehav Rev. 2020;112:39–47. doi:10.1016/j.neubiorev.2020.01.034
28. Su WJ, Zhang Y, Chen Y, et al. NLRP3 gene knockout blocks NF-κB and MAPK signaling pathway in CUMS-induced depression mouse model. Behav Brain Res. 2017;322(Pt A):1–8. doi:10.1016/j.bbr.2017.01.018
29. Liu W, Xu Y, Lu J, Zhang Y, Sheng H, Ni X. Swimming exercise ameliorates depression-like behaviors induced by prenatal exposure to glucocorticoids in rats. Neurosci Lett. 2012;524(2):119–123. doi:10.1016/j.neulet.2012.07.011
30. Liu W, Xue X, Xia J, Liu J, Qi Z. Swimming exercise reverses CUMS-induced changes in depression-like behaviors and hippocampal plasticity-related proteins. J Affect Disord. 2018;227:126–135. doi:10.1016/j.jad.2017.10.019
31. Liu W, Liu J, Xia J, et al. Leptin receptor knockout-induced depression-like behaviors and attenuated antidepressant effects of exercise are associated with STAT3/SOCS3 signaling. Brain Behav Immun. 2017;61:297–305. doi:10.1016/j.bbi.2017.01.001
32. Lach G, Schellekens H, Dinan TG, Cryan JF. Anxiety, Depression, and the Microbiome: a Role for Gut Peptides. Neurotherapeutics. 2018;15(1):36–59. doi:10.1007/s13311-017-0585-0
33. Park AJ, Collins J, Blennerhassett PA, et al. Altered colonic function and microbiota profile in a mouse model of chronic depression. Neurogastroenterol Motil. 2013;25(9):733–e575. doi:10.1111/nmo.12153
34. Winter G, Hart RA, Charlesworth RPG, Sharpley CF. Gut microbiome and depression: what we know and what we need to know. Rev Neurosci. 2018;29(6):629–643. doi:10.1515/revneuro-2017-0072
35. Kelly JR, Borre Y. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res. 2016;82:109–118. doi:10.1016/j.jpsychires.2016.07.019
36. Thapa S, Sheu JC, Venkatachalam A, Runge JK, Luna RA, Calarge CA. Gut microbiome in adolescent depression. J Affect Disord. 2021;292:500–507. doi:10.1016/j.jad.2021.05.107
37. Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi:10.1038/nature08821
38. Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sánchez B, Margolles A. Bifidobacteria and Their Health-Promoting Effects. Microbiol Spectr. 2017;5(3):73. doi:10.1128/microbiolspec.BAD-0010-2016
39. Sánchez B, Delgado S, Blanco-Míguez A, Lourenço A, Gueimonde M, Margolles A. Probiotics, gut microbiota, and their influence on host health and disease. Mol Nutr Food Res. 2017;61(1):1246. doi:10.1002/mnfr.201600240
40. Murros KE, Huynh VA, Takala TM, Saris PEJ. Desulfovibrio Bacteria Are Associated With Parkinson’s Disease. Front Cell Infect Microbiol. 2021;11:652617. doi:10.3389/fcimb.2021.652617
41. Caspi R, Billington R, Keseler IM, et al. The MetaCyc database of metabolic pathways and enzymes - a 2019 update. Nucleic Acids Res. 2020;48(D1):D445–d453. doi:10.1093/nar/gkz862
42. Liu Z, Li L, Ma S, et al. High-Dietary Fiber Intake Alleviates Antenatal Obesity-Induced Postpartum Depression: roles of Gut Microbiota and Microbial Metabolite Short-chain Fatty Acid Involved. J Agric Food Chem. 2020;68(47):13697–13710. doi:10.1021/acs.jafc.0c04290
43. Hassan AM, Mancano G, Kashofer K, et al. High-fat diet induces depression-like behaviour in mice associated with changes in microbiome, neuropeptide Y, and brain metabolome. Nutr Neurosci. 2019;22(12):877–893. doi:10.1080/1028415X.2018.1465713
44. Zhang T, Du Y, Liu X, et al. Study on antidepressant-like effect of protoilludane sesquiterpenoid aromatic esters from Armillaria Mellea. Nat Prod Res. 2021;35(6):1042–1045. doi:10.1080/14786419.2019.1614577
45. Lu X, Ce Q, Jin L, et al. Deoiled sunflower seeds ameliorate depression by promoting the production of monoamine neurotransmitters and inhibiting oxidative stress. Food Funct. 2021;12(2):573–586. doi:10.1039/D0FO01978J
46. Hare BD, Duman RS. Prefrontal cortex circuits in depression and anxiety: contribution of discrete neuronal populations and target regions. Mol Psychiatry. 2020;25(11):2742–2758. doi:10.1038/s41380-020-0685-9
47. Williams LM. Precision psychiatry: a neural circuit taxonomy for depression and anxiety. Lancet Psychiatry. 2016;3(5):472–480. doi:10.1016/S2215-0366(15)00579-9
48. Li XL, Yuan YG, Xu H, et al. Changed Synaptic Plasticity in Neural Circuits of Depressive-Like and Escitalopram-Treated Rats. Int J Neuropsychopharmacol. 2015;18(10):pyv046. doi:10.1093/ijnp/pyv046
49. Kraus C, Klöbl M, Tik M, et al. The pulvinar nucleus and antidepressant treatment: dynamic modeling of antidepressant response and remission with ultra-high field functional MRI. Mol Psychiatry. 2019;24(5):746–756. doi:10.1038/s41380-017-0009-x
50. Chou XL, Fang Q, Yan L, et al. Contextual and cross-modality modulation of auditory cortical processing through pulvinar mediated suppression. Elife. 2020;1:9.
51. Fang Q, Chou XL, Peng B, et al. Circuit via Retino-Colliculo-Pulvinar Pathway Enhances Feature Selectivity in Visual Cortex through Surround Suppression. Neuron. 2020;105(2):355–369.e356. doi:10.1016/j.neuron.2019.10.027
52. Dinan TG, Cryan JF. Gut instincts: microbiota as a key regulator of brain development, ageing and neurodegeneration. J Physiol. 2017;595(2):489–503. doi:10.1113/JP273106
53. De Vadder F, Kovatcheva-Datchary P, Goncalves D, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1–2):84–96. doi:10.1016/j.cell.2013.12.016
54. Muller PA, Schneeberger M, Matheis F, et al. Microbiota modulate sympathetic neurons via a gut-brain circuit. Nature. 2020;583(7816):441–446. doi:10.1038/s41586-020-2474-7
55. Yu KB, Hsiao EY. Roles for the gut microbiota in regulating neuronal feeding circuits. J Clin Invest. 2021;131(10):234. doi:10.1172/JCI143772
56. Wu WL, Adame MD, Liou CW, et al. Microbiota regulate social behaviour via stress response neurons in the brain. Nature. 2021;595(7867):409–414. doi:10.1038/s41586-021-03669-y
57. Li H, Wang P, Huang L, Li P, Zhang D. Effects of regulating gut microbiota on the serotonin metabolism in the chronic unpredictable mild stress rat model. Neurogastroenterol Motil. 2019;31(10):e13677. doi:10.1111/nmo.13677
58. Prabhu VV, Nguyen TB, Cui Y, et al. Metabolite signature associated with stress susceptibility in socially defeated mice. Brain Res. 2019;1708:171–180. doi:10.1016/j.brainres.2018.12.020
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.