Back to Journals » Journal of Inflammation Research » Volume 14
Susceptibility-Related Cytokine Panel for Prediction of Polygonum multiflorum-Induced Hepatotoxicity in Humans
Authors Tu C, Niu M, Wei AW, Tang JF, Zhang L, Jing J, Xiao XH, Wang JB
Received 5 January 2021
Accepted for publication 13 February 2021
Published 4 March 2021 Volume 2021:14 Pages 645—655
DOI https://doi.org/10.2147/JIR.S299892
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Can Tu,1,2 Ming Niu,2 Ai-Wu Wei,3 Jin-Fa Tang,3 Le Zhang,2 Jing Jing,2 Xiao-He Xiao,2 Jia-Bo Wang2,4
1Beijing Research Institute of Chinese Medicine, Beijing University of Chinese Medicine, Beijing, 100029, People’s Republic of China; 2China Military Institute of Chinese Medicine, Fifth Medical Center of Chinese PLA General Hospital, Beijing, 100039, People’s Republic of China; 3The First Affiliated Hospital of Henan University of Chinese Medicine, Zhengzhou, 450000, People’s Republic of China; 4School of Traditional Chinese Medicine, Capital Medical University, Beijing, 100069, People’s Republic of China
Correspondence: Jia-Bo Wang
School of Traditional Chinese Medicine, Capital Medical University, Beijing, 100069, People’s Republic of China
Email [email protected]
Xiao-He Xiao
China Military Institute of Chinese Medicine, Fifth Medical Center of Chinese PLA General Hospital, Beijing, 100039, People’s Republic of China
Email [email protected]
Background: Drug-induced liver injury is a common adverse effect in clinical practice, with severe cases resulting in liver failure and even death. Identification and prediction of individuals susceptible to idiosyncratic DILI continues to remain a challenge.
Methods: In this study, we report that cytokines in human serum can be used to identify and predict individuals susceptible to Polygonum multiflorum-induced DILI (PM-DILI) in retrospective and prospective cohort studies.
Findings: In the retrospective pilot study, we compared serum cytokine expression profiles of the PM-DILI group (n=10) and the PM-Tolerant group (n=12) and found 10 cytokines with significant differences. In the replication cohort study, differences in the 10 cytokines between PM-DILI (n =11) and PM-Tolerant (n=13) groups were verified. Among them, 6 cytokines showed no significant differences at two time points, including liver injury and recovery stage of PM-DILI, suggesting that these 6 cytokines have no correlation with PM-DILI, however, they may be related to susceptibility. Furthermore, all the retrospective cohorts were combined, and a PM-DILI susceptibility prediction model was built by screening the 6 cytokines. The combination of (TNF-α and CCL-2) or VEGF showed the highest sensitivity and specificity. Finally, the efficacy of the above 3 cytokine combination models in predicting PM-DILI-susceptible individuals was verified before PM exposure in another independent prospective cohort (n=24), with sensitivity and specificity of 66.7% and 83.3%, respectively.
Conclusion: This proof-of-concept study demonstrates that the serum cytokine combination reflecting dysimmunity could be used as a new method to predict PM-DILI, thus providing a new perspective for improving the clinical management of IDILI.
Keywords: drug-induced liver injury, Polygonum multiflorum, cytokine, prediction, susceptible individual
Introduction
Drug-induced liver injury (DILI) is a common cause for serious adverse drug reactions in clinical practice.1 Hepatotoxicity is also a major cause of drug development failure, drug withdrawals, and post-marketing warnings.2,3 DILI includes direct, indirect, and idiosyncratic types.4 Idiosyncratic DILI (IDILI) is usually considered as unpredictable based on dose and pharmacological action.5 Even if drugs that cause IDILI do not result in liver injury in the general population, the risk of liver injury rises remarkably in a small number of susceptible individuals after drug exposure, causing liver failure or death in severe cases. IDILI poses critical challenges for drug safety management,6 as it is difficult to predict who is a susceptible individual, even if the hepatotoxic potential of a drug is known.
More progress has been made in the study of IDILI mechanisms in recent years. Host gene polymorphism-related hypotheses have been considered important and are widely accepted and referenced to explain the predisposition of IDILI. Several studies show that human leukocyte antigens (HLA) are associated with IDILI susceptibility to some drugs (eg, HLA-B*57:01 with flucloxacillin; HLA-B*35:02 with minocycline; HLA-DRB1*16:01-DQB1*05:02 with flupirtine).7–9 However, not all patients with susceptible genes have IDILI after taking drugs. Besides the HLA gene, the previous study have found that the IL-10 and IL-4 polymorphisms are associated with susceptibility to diclofenac-induced liver injury.10 Carrying a specific HLA genotype represents an upstream risk factor for the occurrence of IDILI in susceptible individuals, while dysimmunity-related hypotheses (such as cytokines) reflect the downstream risk factors, which are also valuable for identifying and predicting IDILI-susceptible individuals.
In recent studies, some dysimmunity-related IDILI animal models, Yokoi groups showed that the cytokine IL-17 was involved in halothane and α-naphthylisothiocyanate (ANIT)-induced liver injury,11,12 and the Th2-mediated cytokine, IL-4, was involved in dicloxacillin and methimazole-induced live injury.13,14 Roth and colleagues reported that IDILI animal models based on inflammatory stress hypothesis have been developed for several drugs, both IFN-γ and TNF-α were critical for the hepato-pathogenesis including Trovafloxacin, sulindac, amiodarone, and others.15–17 These findings suggested the importance of cytokines in IDILI susceptibility, beyond gene polymorphism. However, these animal models are different than the real-life scenario in humans and therefore may not meet the unique demand of screening susceptible individuals in clinical application, although they are invaluable for studying the mechanisms of IDILI.18 Clinical practice reveals that immunologically tolerant antitumor drugs are more likely to result in liver injury. There is insufficient data from well-designed studies on humans to support the host dysimmunity hypothesis of IDILI. Moreover, there is a lack of predictive biomarker (eg, cytokines) to identify individuals susceptible to IDILI, which restricts our ability to tackle risks of known hepatotoxic drugs in clinical practice.
Polygonum multiflorum Thunb. (PM), a well-known and commonly used traditional Chinese medicine (TCM) and herbal dietary supplement (HDS) for nourishing the kidney and liver, has aroused critical concern for its reported potential DILI.19,20 In our previous research, we demonstrated that PM-DILI occurs only in a small number of individuals who consume the drug, presenting typical characteristics of IDILI.21 Genomic studies have shown that the HLA-B*35:01 allele is a genetic biomarker for predicting PM-DILI in humans.22,23 However, only about 50% of patients with PM-DILI carry the gene mutation, which means that there are other factors and biomarkers that can predict susceptibility in individuals without gene mutation. Thus, PM is a good case to explore multi-factor driven susceptibility to IDILI.
Based on the host dysimmunity hypothesis of IDILI and evidence of animal models, there are theoretical possibilities of finding immunological biomarkers to characterize susceptible individuals. In this study, we demonstrated and verified in real clinical cases that differences in serum immune factors can predict IDILI susceptibility in individuals. This study is also the first clinical case study supporting the host dysimmunity hypothesis of IDILI.
Materials and Methods
This study protocol was conducted in compliance with the ethical principles of the Declaration of Helsinki and was approved by the Medical Ethics Committee, Fifth Medical Centre of Chinese PLA General Hospital (Protocol No. 2017056D). This study included retrospective and prospective cohorts. All participants self-reported their ethnicity as Han Chinese and provided written informed consent.
Retrospective Cohort
The retrospective cohort consisting of PM-DILI patients was collected from the Fifth Medical Centre of Chinese PLA General Hospital (Beijing, China). PM-DILI is defined as the clinical diagnosis of DILI attributed to PM with high probability (RUCAM causality score > 6), based on ACG Clinical Guideline: The Diagnosis and Management of Idiosyncratic Drug-induced Liver Injury, 2014.2,24 Any polypharmacy that may lead to competitive causality of PM was excluded. Twenty-one cases diagnosed as PM-DILI were selected from the retrospective cohort (Figure 1A). Among them, 10 cases were serum specimens collected at the injury stage (no serum specimen at the recovery stage), which was defined as subset 1. Eleven cases included both serum specimens at the injury stage (subset 2) and corresponding serum specimens at the liver function recovery stage (subset 3).
Prospective Cohort
The prospective cohort was collected from the First Affiliated Hospital, Henan University of Traditional Chinese Medicine (Zhengzhou, China). Information about this cohort has been previously published.23,25 Briefly, the outpatients were suitable for PM treatment as prescribed by TCM. The PM dosage (10~15 g) was prescribed by doctor of TCM according to the patient’s condition, and was within the range specified in the 2015 edition of “Chinese Pharmacopoeia”. The patients were prescribed a PM decoction for 4 weeks, which continued for up to 12 weeks. All patients underwent blood collection before or after PM exposure. Liver function was regularly monitored every two weeks to pay attention to timely withdrawal. Patients with underlying liver diseases were excluded. Biochemical indexes of liver functions were all normal before taking PM.
Six patients had an abnormality in biochemical indexes of liver functions at the time of monitoring, who were defined as potential PM-susceptible patients (subset 4). The potential patients susceptible to PM-DILI are defined as those whose liver biochemistry manifested abnormality during digestion (4 weeks) and subsequent prospective monitoring course (12 weeks). Liver biochemistry abnormality is defined as: i) ALT/AST level ≥ 2× ULN with or without TBiL level ≥ 2× ULN or ii) ALP level ≥ 2× ULN after the administration of PM, diagnosed with asymptomatic transaminase elevation. Based on biochemistry criteria of DILI diagnosis. Potential PM-susceptible patients did not meet the criteria for liver injury but manifested potential predisposition to liver injury. To protect the safety of participants, all patients diagnosed with asymptomatic transaminase elevation immediately ceased the PM exposure.
The other patients, who did not show any liver injury during the monitoring period after PM administration, were defined as PM-tolerant patients. Within them, 43 patients with enough serum samples (collected before they took PM) were involved in this study. The nested case–control design (matched ratio, 1:3) was applied to match those six potential PM-susceptible patients (subset 4) with eighteen PM-tolerant patients (subset 5). The remaining 25 cases of PM-tolerant patients were randomly divided into 2 groups as the PM-tolerant control groups for the following comparison and validation analyses, namely subset 6 (12 cases) and subset 7 (13 cases). The case collection and subset grouping are shown in Figure 1B.
Study Design
Reasonably designed screening and validation strategies maximize the possible use of sample resources to obtain a verifiable biomarker panel. For this purpose, we designed four comparative studies, including the pilot study, replication study, model establishment study, and model validation study. The comparisons and modelling strategies are shown in Figure 1C.
I. The pilot study was designed to compare subset 1 (PM-DILI patients sampling at injury) and subset 6 (PM-tolerant patients sampling at pre-dosing). Since the PM-DILI patients in subset 1 were susceptible individuals, the differential expressed cytokines between subsets 1 and 6 (tolerant individuals) may indicate susceptibility. Since the samples in subset 1 were collected at injury stage, the differential expressed cytokines between subsets 1 and 6 may also be associated with liver injury. Thus, the pilot study screens out putative biomarkers (ie, 10 cytokines) associated with either susceptibility or liver injury due to PM-DILI.
II. The replication study was designed to further compare putative biomarkers (screened out in the pilot study) within subset 2 (PM-DILI patients sampling at injury), subset 3 (PM-DILI patients sampling at recovery), and subset 7 (PM-tolerant subjects sampling at pre-dosing). Since either subsets 1 and 2 or 6 and 7 are similar, it can be expected that putative biomarkers being screened out in the pilot study will be replicated in the replication study when comparing subsets 2 and 7. Since patients in subsets 2 and 3 are the same patients whose samples were collected at the time of liver injury or at recovery stages, the biomarkers that changed significantly between the two subsets could be considered as liver injury-related biomarkers. By contrast, unchanged biomarkers between subsets 2 and 3 could be considered as candidate biomarkers (ie, 6 cytokines) associated with susceptibility rather than liver injury.
III. In the subsequent model establishment study, we combined samples in the pilot and replication studies and tried to establish a prediction model for individuals susceptible to PM-DILI, based on the candidate biomarkers (screened out in the replication study). The optimal prediction model consists of one or more biomarkers based on the positive and negative prediction rates of the model (ie, a panel of 3 cytokines).
IV. In the final model validation study, we tested the performance of the prediction model (screened out in the model establishment study) in the prospective cohort samples, independent of the former samples used in model establishment. In order to be truly predictive, the changes in plasma cytokines were measured before PM exposure. The sensitivity, specificity, positive, and negative prediction rates of the prediction model were assessed.
Determination of Serum Cytokines
All serum samples retrieved from the biobank were thawed before assay by the Luminex liquid phase chip technology to determine the 20 cytokines, including chemokine (C-C motif) ligands (CCL2, CCL3, and CCL11) chemokine (C-X3-C motif) ligand 1 (CX3CL1), C-X-C motif chemokine ligands (CXCL1 and CXCL10), epidermal growth factor (EGF), granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony stimulating factor (GM-CSF), interferon-γ (IFN-γ), interleukins (IL-1α, IL-1β, IL-2, IL-5, IL-6, IL-10, IL-12, and IL-17α), tumor necrosis factor-α (TNF-α), and vascular endothelial growth factor (VEGF). The multiplex immunoassay kit used was provided by eBioscience, USA (Lot: 132,299,102). Samples were diluted and incubated with magnetic beads conjugated to capture antibodies. After washing, samples were incubated with diluted detection antibody. Streptavidin–phycoerythrin was then added for incubation, and finally data was acquired using a MAGPIX instrument.
Statistical Analysis
Experimental data was analyzed with the SPSS software program (version 18.0, Chicago, IL, USA) and expressed as mean ± SD. One-way analysis of variance (ANOVA) and t-test were used for statistical analysis of the results. A P value <0.05 was statistically significant. A logistic regression model was used to investigate the association of cytokine alleles with PM-DILI. Odds ratio (OR) was calculated using a 2×2 contingency table, which added 0.05 to all fields to accommodate possible zero counts.
Results
Clinical Characteristics
In the 21 cases of PM-DILI in the retrospective cohort, there were more female patients than male, the median age of incidence of PM-DILI was 45 years old. The majority of PM-DILI cases was hepatocellular injury pattern (19/21, 90.4%); and the other two cases were cholestatic and mixed types respectively. All patients had a good prognosis after drug withdrawal, with 21 (13, 25) days as the median time of hospital stay. The biochemical indexes of liver functions returned to normal within 6 weeks, with the longest recovery time of 4 months (Table S1).
A total of 49 female patients in the prospective cohort were eligible for the study, including 6 potential PM-susceptible patients and 43 PM-tolerant patients. There was no significant difference between the potential PM-susceptible group and the PM-tolerant group in age, gender, dose, duration of medication, etc. (P>0.05). Biochemical indexes of liver injury (ALT, AST, ALP, and TBil) before PM administration were not significantly different between the two groups (P>0.05). Compared to the PM-tolerant group, ALT and AST were significantly increased in the PM-susceptible group after PM administration (P<0.01). There was no significant difference in dose and duration of medication between the potential PM-susceptible group and the PM-tolerant group. (Table 1).
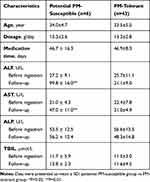 |
Table 1 Clinical Characteristics of Patients from the Prospective Cohort Administered with PM |
Pilot Study for Liver Injury- or Susceptibility-Related Cytokines
According to the pilot study design (see details in Study Design section), subsets 1 (PM-DILI/at injury) and 6 (PM-tolerant/at pre-dosing) were compared, and those cytokines with significant differences between the two groups can be considered as putative biomarkers related to either liver injury or susceptibility. Subset 1 consisted of blood samples from PM-DILI patients collected at the injury stage, with mean serum values of 27 ULN ALT and 21 ULN AST (Figure 2A). Subset 6 comprised of blood samples from PM-tolerant patients collected before they were administered PM, whose serum ALT and AST were within the normal range. There was a significant difference in transaminases between subsets 1 and 6 (P<0.01). Further multivariate statistical analysis was performed with 20 cytokines as variables. Subsets 1 (PM-DILI/at injury) and 6 (PM-tolerant/at pre-dosing) were easily distinguished in either principal component analysis (PCA) or OPLS-DA analysis (Figure 2B and C). Among them, a total of 10 cytokines showed significant differences between the two groups, including 9 increased cytokines (CCL2, CCL11, CXCL1, CXCL10, EGF, IFN-γ, TNF-α, IL-10, and VEGF) and 1 decreased cytokine (CX3CL1) (Figure 2D, Table S2). These 10 cytokines were thus assumed as putative biomarkers associated to either susceptibility or liver injury of PM-DILI.
Replication Study for Susceptibility-Related Cytokines
Based on the design of the replication study, subset 2 (PM-DILI patients sampling at injury), subset 3 (PM-DILI patients sampling at recovery), and subset 7 (PM-tolerant subjects sampling at pre-dosing) were compared based on 10 putative biomarkers (screened out in pilot study). We found that 6 cytokines, including CCL2, CCL11, CXCL10, CX3CL1, TNF-α, and VEGF, showed no differences between subsets 2 (PM-DILI patients sampling at injury) and 3 (PM-DILI patients sampling at recovery), but manifested significant differences between subset 7 (PM-tolerant subjects sampling at pre-dosing) and each of the other two subsets (Figure 3A, Table S3). Considering subsets 2 and 3 were all PM-DILI patients and were obviously susceptible, we speculate that these 6 cytokines are most likely to be susceptibility-related biomarkers of PM-DILI. We then performed PCA analysis based on these 6 susceptibility-related cytokines. The results showed that the PM-DILI samples irrespective of sampling stages (subset 2 and subset 3, ie, the susceptible ones), scattered in nearly the same zones, while the PM-tolerant samples were distributed in a different area (Figure 3B). These results further support that the 6 identified cytokines may be the susceptibility-related biomarkers that can differentiate PM-DILI-susceptible individuals from tolerant ones.
We found the other 4 cytokines, including CXCL1, EGF, IFN-γ, and IL-10, showed significant differences between subset 2 (PM-DILI patients sampling at injury) and subset 3 (PM-DILI patients sampling at recovery) (Figure 3C, Table S3), which means these 4 cytokines related to liver injury. As expected, the following PCA analysis based on these 4 cytokines showed a clear differentiation between subsets 2 and 3; however, subset 3 (PM-DILI patients sampling at recovery) and subset 7 (PM-tolerant subjects sampling at pre-dosing) could not be differentiated (Figure 3D).
Considering these results, the 6 cytokines, including CCL2, CCL11, CXCL10, CX3CL1, TNF-α, and VEGF, were screened out as the candidate biomarkers related to susceptibility to PM-DILI.
Establishing a Prediction Model for PM-DILI-Susceptible Individuals
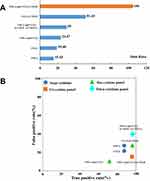
Based on the six susceptibility-related cytokines, we created a prediction model to identify PM-DILI-susceptible individuals. The samples from the pilot and replication studies were combined to assess the performance of the models that were screened by each single cytokine or multi-cytokine panel. The results of single-cytokine models showed that TNF-α (OR = 19.0) or CCL2 (MCP-1, OR = 15.4) had high OR values to differentiate susceptible patients from tolerant ones, while VEGF, CX3CL1 (Fractalkine), CCL11 (Eotaxin), and CXCL10 (IP-10) had lower OR values (>3 and <6). The cutoff values of each cytokine are depicted in Table S4. The positive prediction rates of TNF-α- or CCL2-based single cytokine model exceeded 80% and the negative prediction rates were less than 30%.
We further screened all the combinations of dual cytokine panels. The results showed that there were six kinds of dual cytokine models that had higher OR values than the highest OR value of single cytokine model, which were “CCL2 or VEGF” (OR = 51.4), “TNF-α or VEGF” (OR = 42.5), “TNF-α or CXCL10” (OR = 35.6), “CCL2 or CXCL10” (OR = 35.6), “TNF-α or CCL2” (OR = 30.0), and “TNF-α and CCL2” (OR = 23.5) (Table S5). Except for “TNF-α and CCL2”, the other five dual cytokine models all achieved higher positive prediction rates (90%) than the single cytokine models. However, the negative prediction rates did not improve.
Similarly, we screened all the combinations of tri-cytokine panels. The results showed that only two tri-cytokine combinations, “(TNF-α and CCL2) or VEGF” (OR = 105. 0) and “(TNF-α and CCL2) or CXCL10” (OR = 63.3) (Table S6), had higher OR values than the dual cytokine models. The tri-cytokine model of “(TNF-α and CCL2) or VEGF” achieved best positive prediction rate (95.3%) and low negative prediction rate (16.0%). However, when we tried to construct models exceeding three cytokines, there was no further performance improvement as compared to the tri-cytokine model.
Typical cytokine models have been summarized in Figure 4A and B. It can be seen that the tri-cytokine panel “(TNF-α and CCL2) or VEGF” is the best model to predict PM-DILI-susceptible individuals, with highest OR value and best true/false positive rates.
Validation of Tri-Cytokine Panel-Based Prediction Model
Nested case–control paired prospective cohorts were used to assess the tri-cytokine panel-based prediction model. This helped to explore the value of serum cytokine panel for prediction of individuals susceptible to PM-DILI before drug exposure. All the enrolled patients had normal ALT, AST, TBiL, and ALP levels before they were administered PM (Table 2). After 4 weeks of monitoring, 6 patients (potential PM-susceptible individuals) showed a significant increase in ALT and AST levels (≥2 × ULN) compared to the other nested paired subjects (PM-tolerant individuals) with normal ALT and AST levels. By using the tri-cytokine panel-based prediction model (“TNF-α and CCL2 or VEGF”), good performance (sensitivity 66.7% and specificity 83.3%) was achieved before PM exposure in the prospective cohort. Positive and negative predictive value of the model were 57.1% and 88.2%, respectively.
 |
Table 2 Prediction Performance of the Tri-Cytokine Model |
Discussion
Obtaining high-quality biological specimens of susceptibility-related cases is challenging due to the low incidence rate and unpredictability of IDILI. Therefore, it is essential to use collected PM-DILI cases in clinical practice and prospectively monitor susceptible cases. In this study, the cytokine panel (TNF-α and CCL2) or VEGF was identified as a potential high-risk, dysimmunity causing factor for PM-DILI that is specific for PM. Four comparative studies were designed to assess the association between PM-DILI and the cytokine panel. Our study revealed that patients with PM-DILI returned to normal levels of serum liver function indexes after recovery, however, compared with PM-tolerant patients, the PM-DILI recovery group showed significant differences between six cytokines, including CCL2, CCL11, CXCL10, CX3CL1, TNF-α, and VEGF. The results suggest that these six highly expressed cytokines may not be related to PM-DILI, but may highly correlate with susceptibility. In conclusion, the serum transaminase level in PM-DILI patients indicated that substantive liver damage had been repaired, and that the immune cytokines of some patients exhibited higher expression levels, indicating that the immune microenvironment of susceptible individuals was still under immune stress. Furthermore, it suggests that immune susceptibility factors may be isolated from the liver injury process. The immunity of population susceptible to liver injury was in a stimulated pro-inflammatory state, accompanied with the high expression state of most chemokines.
On this basis, we verified the clinical predictive efficacy of selected cytokines with a higher correlation of PM-DILI susceptibility of patients. We combined samples from two independent retrospective studies to build a prediction model of PM-DILI-susceptible population (21 PM-DILI patients, 25 PM-tolerant patients). Through univariate and multi-factor combination studies, we found that the cytokine combination, TNF-α, and CCL2 or VEGF, could be used as a biomarker to predict the PM-DILI-susceptible population with the highest predictive efficacy. In total, within this PM-DILI prediction model, the risk of liver injury for cytokine combination (TNF-α and CCL2) or VEGF was 18 times higher than that of non-high expression group. In total, 95.24% of PM-DILI patients (20/21) expressed a positive cytokine panel (TNF-α and CCL2) or VEGF, whereas 16% of PM-tolerant patients (4/25) expressed a similar positive cytokine panel. In the model establishment study, compared to the expression of negative cases, individuals expressing positive cytokine panel (TNF-α and CCL2) or VEGF had a several-fold increased risk of developing PM-related liver injury. Therefore, we confirm that individuals expressing positive cytokine panel (TNF-α and CCL2) or VEGF are at a higher risk of developing DILI when treated with herbal remedies containing PM. Finally, in a prospective cohort study, 4/7 (57.1%) positive cytokines panel cases “(TNF-α and CCL2) or VEGF” and 2/17 (11.8%) negative cytokines panel cases developed 2-fold elevation of alanine aminotransferase, confirming that individuals of cytokines panel “(TNF-α and CCL2) or VEGF” are at a higher risk of developing DILI when exposed to PM.
The current theory suggests that the majority of idiosyncratic drug reaction have evidence to support immune involvement.3 Recent progress in clinical and basic studies has implied that PM-DILI is more likely to be immune-mediated IDILI.26,27 Clinical liver biopsies revealed that some PM-DILI patients had marked mixed inflammatory cell infiltration and Kupffer cell activation.23 Changes in levels of circulating cytokines and chemokines have been proposed as possible biomarkers of DILI. A PM-DILI model based on innate immune-mediated immunological stress was established in our previous research, cotreatment with nontoxic dose LPS and PM (the clinically 2 times equivalent dose) caused liver injury accompanied by the activation and infiltration of inflammatory cells (such as Kupffer cells) and the release of proinflammatory cytokines and chemokines (TNF-α, IL-1β and CCL2, etc.).20,28 In deeds, several groups have reported that the cytokines were involved in IDILI pathogenesis of drugs in animal models (eg, IL-17 with halothane and ANIT,13 IL-4 with dicloxacillin and methimazole,12 TNF-α combined IFN-γ with trovafloxacin15). In this paper, the results revealed for the first that the cytokines have a good predictive in potential PM-DILI susceptible individuals in humans. Despite the small sample size, prospective cohort study showed that the optimal tri-cytokine panel “TNF-α and CCL2 or VEGF” had high sensitivity for predicting PM-DILI. These results suggested the cytokines, that possibly be of predictive value, could provide extremely useful information to understand the mechanisms of PM-DILI or other IDILI drugs.
Evidence in humans and animals suggests that drugs can be bioactivated to reactive metabolites that can initiate an adaptive immune response. Alternatively, initiation of an innate immune response through TLR activation can prompt a hepatotoxic interaction with drugs.16 Both of these immune responses result in the generation of mediators, such as TNF-a and IFN-γ. Meanwhile, Chemokines (CCL-2 and CXCL2), that induced the infiltration of Kupffer cells and neutrophils followed by necrosis, might be involved in the dicloxacillin-induced liver injury.13 CCL-2 was increased in acetaminophen-induced liver injury.29 In this paper, CCL-2 was demonstrated to be involved in the PM-DILI. In addition, the cytokines (such as IFN-γ) might induce some IDILI-associated drugs inhibit hepatocyte proliferation, thereby impairing the ability of the liver to adaptation.30,31 In brief, the host-dependent mechanisms associated with immune factors, including individual immune stress status (eg, cytokine) and immunogenetic variations, contribute to the risk of PM-DILI.
Keeping in mind patient safety, PM was not administrated for a long period of time (about 30 days) in the prospective cohort study. Most PM-DILI occurred during 1 to 2 months. Therefore, the current data may underestimate the occurrence of PM-DILI, which may provide an explanation for the 3 patients with a positive cytokine panel (TNF-α and CCL2) or VEGF who took PM but did not show elevated ALT in the cohort. By design, the study involved one ethnic group, one agent, and focused on dysimmunity factors only. Therefore, the results must be viewed within such a predefined context. In addition, a small-sample-size prospective cohort study brings uncertainty and variation to the prediction of PM-DILI-susceptible individuals, so we need a larger, independent prospective cohort of PM-DILI patients to further verify the findings in this study. According to our current study results, positive cytokine panels “(TNF-α and CCL2) or VEGF” are necessary but insufficient for predicting PM-DILI-susceptible individuals. However, the positive cytokine panel can be used as a supplementary method for the diagnosis of PM-DILI and a prospective screening tool for predicting the risk of PM-DILI.
Conclusions
In summary, our study reported the panel of serum cytokines (TNF-α and CCL2) or VEGF as potential biomarkers of PM-DILI, thus providing insights into the mechanism of PM-DILI. This is the first clinical case study confirming that serum immune factor differences can predict IDILI-susceptible individuals and supports the susceptibility hypothesis of the host dysimmunity mediating IDILI. Prospective cohorts with a larger number of subjects are consequently needed to investigate the potential clinical utility of cytokine markers in identifying individuals susceptible to PM-DILI, and to elucidate the molecular mechanisms of PM-DILI.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was financially supported by the National Natural Science Foundation of China (grant numbers No. 82074112, 81630100 and 81721002). Project of China PLA General Hospital (2019-JQPY-003 and 2019MBD-023).
Disclosure
The authors declare no conflict of interests.
References
1. Navarro VJ, Senior JR. Drug-related hepatotoxicity. N Engl J Med. 2006;354(7):731–739. doi:10.1056/NEJMra052270
2. Chalasani NP, Hayashi PH, Bonkovsky HL, Navarro VJ, Lee WM, Fontana RJ. ACG clinical guideline: the diagnosis and management of idiosyncratic drug-induced liver injury. Am J Gastroenterol. 2014;109(7):
3. Chen M, Suzuki A, Borlak J, Andrade RJ, Lucena MI. Drug-induced liver injury: interactions between drug properties and host factors. J Hepatol. 2015;63(2):503–514. doi:10.1016/j.jhep.2015.04.016
4. Hoofnagle JH, Bjornsson ES. Drug-induced liver injury - types and phenotypes. N Engl J Med. 2019;381(3):264–273. doi:10.1056/NEJMra1816149
5. Andrade RJ, Chalasani N, Björnsson ES, et al. Drug-induced liver injury. Nat Rev Dis Primers. 2019;5(1):58. doi:10.1038/s41572-019-0105-0
6. Björnsson E. Review article: drug-induced liver injury in clinical practice. Aliment Pharmacol Ther. 2010;32(1):3–13. doi:10.1111/j.1365-2036.2010.04320.x
7. Urban TJ, Nicoletti P, Chalasani N, et al. Minocycline hepatotoxicity: clinical characterization and identification of HLA-B*35:02 as a risk factor. J Hepatol. 2017;67(1):137–144. doi:10.1016/j.jhep.2017.03.010
8. Nicoletti P, Werk AN, Sawle A, et al. HLA-DRB1*16: 01-DQB1*05: 02 is a novel genetic risk factor for flupirtine-induced liver injury. Pharmacogenet Genomics. 2016;26(5):218–224. doi:10.1097/FPC.0000000000000209
9. Daly AK, Donaldson PT, Bhatnagar P, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet. 2009;41(7):816–819. doi:10.1038/ng.379
10. Aithal GP, Ramsay L, Daly AK, et al. Hepatic adducts, circulating antibodies, and cytokine polymorphisms in patients with diclofenac hepatotoxicity. Hepatology. 2004;39(5):1430–1440. doi:10.1002/hep.20205
11. Kobayashi E, Kobayashi M, Tsuneyama K, Fukami T, Nakajima M, Yokoi T. Halothane-induced liver injury is mediated by interleukin-17 in mice. Toxicol Sci. 2009;111(2):302–310. doi:10.1093/toxsci/kfp165
12. Kobayashi M, Higuchi S, Mizuno K, et al. Interleukin-17 is involved in alpha-naphthylisothiocyanate-induced liver injury in mice. Toxicology. 2010;275(1–3):50–57. doi:10.1016/j.tox.2010.05.011
13. Higuchi S, Kobayashi M, Yoshikawa Y, et al. IL-4 mediates dicloxacillin-induced liver injury in mice. Toxicol Lett. 2011;200(3):139–145. doi:10.1016/j.toxlet.2010.11.006
14. Kobayashi M, Higuchi S, Ide M, et al. Th2 cytokine-mediated methimazole-induced acute liver injury in mice. J Appl Toxicol. 2012;32(10):823–833. doi:10.1002/jat.2731
15. Poulsen KL, Olivero-Verbel J, Beggs KM, Ganey PE, Roth RA. Trovafloxacin enhances lipopolysaccharide-stimulated production of tumor necrosis factor-α by macrophages: role of the DNA damage response. J Pharmacol Exp Ther. 2014;350(1):164–170. doi:10.1124/jpet.114.214189
16. Roth RA, Maiuri AR, Ganey PE. Idiosyncratic drug-induced liver injury: is drug-cytokine interaction the linchpin? J Pharmacol Exp Ther. 2017;360(2):461–470. doi:10.1124/jpet.116.237578
17. Roth RA, Ganey PE. Animal models of idiosyncratic drug-induced liver injury–current status. Crit Rev Toxicol. 2011;41(9):723–739. doi:10.3109/10408444.2011.575765
18. Jennings JJ, Mandaliya R, Nakshabandi A, Lewis JH. Hepatotoxicity induced by immune checkpoint inhibitors: a comprehensive review including current and alternative management strategies. Expert Opin Drug Metab Toxicol. 2019;15(3):231–244. doi:10.1080/17425255.2019.1574744
19. Wang J, Ma Z, Niu M, et al. Evidence chain-based causality identification in herb-induced liver injury: exemplification of a well-known liver-restorative herb Polygonum multiflorum. Front Med. 2015;9(4):457–467. doi:10.1007/s11684-015-0417-8
20. Tu C, He Q, Li CY, et al. Susceptibility-related factor and biomarkers of dietary supplement polygonum multiflorum-induced liver injury in rats. Front Pharmacol. 2019;10:335. doi:10.3389/fphar.2019.00335
21. He LZ, Yin P, Meng YK, et al. Immunological synergistic mechanisms of trans-/cis-stilbene glycosides in Heshouwu-related idiosyncratic liver injury. Sci Bull. 2017;62(11):748–751. doi:10.1016/j.scib.2017.04.020
22. Yang WN, Pang LL, Zhou JY, et al. Single-nucleotide polymorphisms of HLA and Polygonum multiflorum-induced liver injury in the Han Chinese population. World J Gastroenterol. 2020;26(12):1329–1339. doi:10.3748/wjg.v26.i12.1329
23. Li C, Rao T, Chen X, et al. HLA-B*35:01 allele is a potential biomarker for predicting polygonum multiflorum-induced liver injury in humans. Hepatology. 2019;70(1):346–357. doi:10.1002/hep.30660
24. Xiao XH, Li XH, Zhu Y, et al. Guideline for diagnosis and treatment of herb-induced liver injury. Zhongguo Zhong Yao Za Zhi. 2016;07:1165–1172.
25. Zhang L, Niu M, Wei AW, et al. Risk profiling using metabolomic characteristics for susceptible individuals of drug-induced liver injury caused by Polygonum multiflorum. Arch Toxicol. 2020;94(1):245–256. doi:10.1007/s00204-019-02595-3
26. Meng YK, Li CY, Li RY, et al. Cis-stilbene glucoside in Polygonum multiflorum induces immunological idiosyncratic hepatotoxicity in LPS-treated rats by suppressing PPAR-gamma. Acta Pharmacol Sin. 2017;38(10):1340–1352. doi:10.1038/aps.2017.32
27. Li CY, He Q, Gao D, et al. Idiosyncratic drug-induced liver injury linked to Polygonum multiflorum: a case study by pharmacognosy. Chin J Integr Med. 2017;23(8):625–630. doi:10.1007/s11655-017-2543-9
28. Li C, Niu M, Bai Z, et al. Screening for main components associated with the idiosyncratic hepatotoxicity of a tonic herb, Polygonum multiflorum. Front Med. 2017;11(2):253–265. doi:10.1007/s11684-017-0508-9
29. Dambach DM, Durham SK, Laskin JD, Laskin DL. Distinct roles of NF-kappaB p50 in the regulation of acetaminophen-induced inflammatory mediator production and hepatotoxicity. Toxicol Appl Pharmacol. 2006;211(2):157–165. doi:10.1016/j.taap.2005.06.024
30. Dong Z, Zhang J, Sun R, Wei H, Tian Z. Impairment of liver regeneration correlates with activated hepatic NKT cells in HBV transgenic mice. Hepatology. 2007;45(6):1400–1412. doi:10.1002/hep.21597
31. Brooling JT, Campbell JS, Mitchell C, Yeoh GC, Fausto N. Differential regulation of rodent hepatocyte and oval cell proliferation by interferon gamma. Hepatology. 2005;41(4):906–915. doi:10.1002/hep.20645
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.