Back to Journals » Cancer Management and Research » Volume 11
Suppression of miR-21-3p enhances TRAIL-mediated apoptosis in liver cancer stem cells by suppressing PI3K/Akt/Bad cascade via regulating PTEN
Authors Zhu Y, Tang H, Zhang L, Gong L, Wu G, Ni J, Tang X
Received 9 August 2018
Accepted for publication 19 October 2018
Published 22 January 2019 Volume 2019:11 Pages 955—968
DOI https://doi.org/10.2147/CMAR.S183328
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Rituraj Purohit
Yingwei Zhu,1,* Hong Tang,2,* Lili Zhang,1 Lei Gong,1 Gaojue Wu,1 Jingbin Ni,1 Xuejun Tang1
1Department of Gastroenterology, The Affiliated Wuxi No.2 People’s Hospital of Nanjing Medical University, Wuxi, Jiangsu 214002, People’s Republic of China; 2Department of Pathology, The Affiliated Wuxi No.2 People’s Hospital of Nanjing Medical University, Wuxi, Jiangsu 214002, People’s Republic of China
*These authors contributed equally to this work
Background: TNF-related apoptosis-inducing ligand (TRAIL) functions as a selective apoptosis-inducing ligand in cancer cells with normal cells remaining unaffected; however, resistance limits its anticancer properties. Cancer stem cells (CSCs) are involved in the treatment of resistant cancer cases including liver cancer (LC). The aim of this study was to look into the approaches for increasing the sensitivity of liver cancer stem cells (LCSCs) toward TRAIL.
Methodology: PLC, HepG2 and Huh7 LC cell lines were used in this study. Quantitative reverse transcription PCR (qRT-PCR) analysis was done for evaluating the expression of miR-21-3b. Fluorescent-activated cell-sorting equipment was used for separation and identification of LCSCs and non-LCSCs. The cells were transfected with RNA along with miR-21-3p mimics, anti- miR-21-3p, miR-NC and the phosphatase and tensin homologue (PTEN) siRNA. MTT assay for cell viability, Luciferase assay for luciferase activity, Western blots for the expression of proteins and flow cytometry for the measurement of ROS and apoptosis, respectively, were carried out. Tumor xenografts nude mice were used for tumor growth in vivo.
Results: We found that miR-21-3p was overexpressed in LCSCs compared to non-LCSCs and that the suppression of miR-21-3p along with anti-miR-21-3p enhanced the sensitivity of LCSCs to TRAIL-mediated apoptosis. We further found that miR-21-3p regulated the expression of PTEN in Huh7-LCSCs directly and that the suppression of miR-21-3p enhanced the levels of PTEN. The study confirmed that inhibition of the PI3K/Akt/Bad signaling pathway was involved in enhancing TRAIL-mediated apoptosis of LC cells.
Conclusion: The study suggested that overexpression of miR-21-3p suppresses the sensitivity to TRAIL in LCSCs. This study concludes that the suppression of miR-21-3p is a potential approach for enhancing the sensitivity of LC cells toward TRAIL by PI3K/Akt/Bad cascade via the miR-21-3p/PTEN axis.
Keywords: TRAIL, liver cancer, miR-21-3p, PI3K, Akt, Bad, PTEN
Introduction
Liver cancer (LC) also known as cancer of hepatic cells is one of the leading malignancies responsible for deaths caused by cancer worldwide.1 In the need of finding the mechanism responsible for tumorigenic transformation of LC, a number of studies have been carried out; however, the studies suggesting specific molecular targets for treating LC still remain insufficient.2 Recent studies have confirmed that cancer is a stem cell disorder. According to the findings, tumors are abnormal growths made up of a group of cells called as cancer stem cells (CSCs), which have the ability to self-renew.3,4
A study recently has confirmed CD-133 as the potential marker of LC stem cells (LCSCs).5 These cells have been identified to possess many important properties such as colony formation, greater ability to form tumors and resistance against chemotherapy.6 All these properties make LCSCs as favorable targets for treating LC.
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) is a member of TNF family cytokine proteins.7 TRAIL plays a crucial role in cancer; it selectively activates apoptosis in tumor cells without affecting the normal cells.8 However, some of the CSCs are resistant to TRAIL-mediated apoptosis, and the mechanism of resistance involved is still not clear.9 This makes urgency to find the involved mechanism and measures to reduce the resistance of LCSCs against TRAIL.
miRNAs are small non-coding RNAs, which act as potential regulators by binding to the 3′-UTR of the target mRNAs.10,11 A number of studies have confirmed miRNAs to play important role in metabolism, proliferation, apoptosis and differentiation of cells. Abnormal expression of miRNAs is associated with various disorders, with cancer being the major one.12,13
miR-21 is one of the mRNAs displaying oncogenic property and is upregulated in most of the cancers.14 miR-21 has the following two strands: miR-21-5p (guide strand) and miR-21-3p (passenger strand). A number of studies have been floated, suggesting tumorigenesis character of miR-21-5p in most of the stages of tumorigenesis such as drug resistance metastasis, angiogenesis, proliferation, metabolism and invasion.15–18 Among the two strands, miR-21-5p is less stable and is preferably selected by target cells, whereas miR-21-3p is more stable and is destroyed by cells.19 Upregulation of miR-21-5p has been reported in ovarian cancer20–23 and has been found to indulge chemoresistance in ovarian, breast and lung cancers.24–26 Reports recently have pointed that both the guide strand (miR-21-5p) and the passenger strand (miR-21-3p) of miR-21 target same number of genes and are responsible for the regulation of the expression of their target genes.27 Besides being responsible to regulate equal number of genes, compared to the guide strand (miR-21-5p), the role of the passenger strand (miR-21-3p) in cancer and resistance studies is not fully elucidated. In a previous study, miR-21-3p was found to function as a tumor suppressor in hepatic cancer, suggesting its therapeutic role;28 however, its role in enhancing TRAIL-mediated apoptosis remains unclear. With the aim to investigate the measures for enhancing the TRAIL-mediated sensitivity of LCSCs, in this study we discovered that the passenger strand (miR-21-3p) was significantly unregulated in LCSCs against the non-LCSCs. Study also suggested that the suppression of miR-21-3p enhanced the sensitivity of LCSCs against TRAIL-mediated apoptosis. Importantly, we established that upregulation of miR-21-3p suppresses the sensitivity of LCSCs to TRAIL.
Materials and methods
Animals
For this study, nude BALB/C mice of either sexes aging 4–5 weeks and weighing 20–25 g were selected, which were obtained from the Department of Gastroenterology, The Affiliated Wuxi No. 2 People’s Hospital of Nanjing Medical University, Wuxi, Jiangsu, People’s Republic of China. The animals were housed in polypropylene cages maintained at 24°C with dark–light cycle of 12 hours. The mice received food and water ad libitum.
Ethical approval and consent to participate
The animal protocol was approved by Animal Care Committee of – The Affiliated Wuxi No.2 People’s Hospital of Nanjing Medical University (Approval number: LC/NMU/2016-17/041A); all the experiments adhered to the draft of “Animal protection law of the People’s Republic of China-2009” for experimental animals.
Cell lines and culture conditions
For this study, we selected PLC, HepG2 and Huh7 that were cancerous and the normal liver cells (L-O2). The cells were collected from The Affiliated Wuxi No.2 People’s Hospital of Nanjing Medical University, People’s Republic of China. The use of cell lines was approved by the hospital institutional ethics committee. The selected cell lines were cultured in DMEM supplemented with FBS (10%) in humid conditions (5%) at 37°C in an incubator. To study the role of an anti-miR-21-3p in LC cell lines in animal models, the selected Huh7 cells with miR-21-3p knockdown were produced by using lentiviral vector having miR-21-3p antisense nucleotide sequence (Cell signaling Technology, Danvers, MA, USA) (5×105 units) to transfect Huh7 cells.
Expression level of miR-21-3p in the selected cell lines
Quantitative reverse transcription PCR (qRT-PCR) analysis was done for studying the expression level of miR-21-3p (5′-CGACAACAGUCUGAAGGCUGUC-3′) in three cancerous and one non-cancerous cell lines using the primer. Briefly, the total RNA was isolated from the selected cell lines with the help of TRI Reagent (Sigma-Aldrich Co., St Louis, MO, USA) by following the manufacturer’s protocol. The SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific, Waltham, MA, USA) was used to perform reverse transcription followed by qRT-PCR using Thermo Scientific DreamTaq Hot Start DNA Polymerase (Thermo Fisher Scientific).
Separation and identification of LCSCs and non-LCSCs
LC cell lines were obtained from American Tissue Culture Collection (ATCC, Manassas, VA, USA) and washed at least two times before further use. The cells were subjected to staining using anti-CD133-FITC (Merck & Co., Inc., Whitehouse Station, NJ, USA) for 15 minutes. The sorting of CD133+ cells into LCSCs and their percentage was done using fluorescent-activated cell-sorting equipment (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Transfection
The cells were transfected using Lipofectamine™ 2000 reagent (Thermo Fisher Scientific) with RNA oligonucleotides (50 pmol/mL) along with miR-21-3p mimics, anti-miR-21-3p, the control RNA oligonucleotides also termed as miR-NC and the small interfering RNA of phosphatase and tensin homologue (PTEN) also called as PTEN siRNA (RiboBio Co., Ltd., Guangzhou, People’s Republic of China).
The MTT assay for cell viability studies
Before transfecting the selected LCSCs and non-LCSCs, the selected cell lines were seeded in 96-well plates at 5×103 cells/well.44 After 24 hours of transfection, the cells were incubated along with various concentrations of TRAIL for next 48 hours. After incubation, the cells along with medium were exposed to MTT (Thermo Fisher Scientific) (20 µL) at a concentration of 5 mg/L for next 4 hours. The cells after 4 hours were subjected to lysis using a lysis buffer (Bio-Rad Laboratories Inc.) for 15 minutes at room temperature. The absorbance at 570 nm was recorded using a microplate reader (Thermo Fisher Scientific) for each cell.
Luciferase reporter assay
The fragment of PTEN 3′-UTR having the seed region of miR-21-3p was cloned for creating pGL3-PTEN, which is the PGL3 reporter vector. Site-directed mutagenesis kit having pGL3-mutant PTEN was used to create the mutant PTEN reporter. The LCSCs were seeded in 96-well plates for Luciferase assay, and the cells were co-transfected with miR-21-3p mimics or inhibitors (50 pmol/mL) along with Renilla luciferase and Firefly luciferase reporters (100 ng/mL) and pRL-TK vector. The luciferase assay was recorded after 48 hours of transfection using the Dual-Luciferase Reporter assay system (Promega Corporation, Fitchburg, WI, USA) by following the manufacturer’s instructions.
Isolation of mitochondria
For finding the release of cytochrome C, nuclear gene DIABLO direct inhibitor of apoptosis-binding protein was used, which has low apoptosis-inducing factor (AIF) and pI in cytoplasm. The mitochondria of LSCSs were separated with the help of mitochondria separating kit (Sigma-Aldrich Co.) by following the manufacturer’s protocol.
Immunoprecipitation
The cells were subjected to lysis (RIPA buffer; Sigma-Aldrich Co.) followed by centrifugation (12,000x g) for 15 minutes for separating cells. The obtained supernatants were incubated with primary antibodies for Bad (Thermo Fisher Scientific) at 4°C for 12 hours. The protein Agarose beads were subjected to incubation for 1.5 hours and were then washed using cold RIPA buffer and boiled in SDS buffer for isolating the co-precipitated proteins.
Immunoblotting studies
The obtained co-precipitated proteins (50 µg) were subjected to separation using SDS-PAGE (10%). The membranes were blocked using non-fat milk (5%) followed by incubation with various specific antibodies (Table 1). The membranes were incubated with secondary horseradish peroxidase-conjugated antibody (Abcam, Cambridge, UK), and enhanced chemiluminescence (ECL) reagent (Thermo Fisher Scientific) was used for detecting signals.
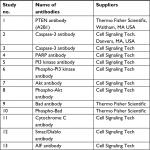  | Table 1 Antibodies used for the study Abbreviations: AIF, apoptosis-inducing factor; PTEN, phosphatase and tensin homologue. |
Measurement of ROS and apoptosis
For measuring ROS and extent of apoptosis, flow cytometry study was done. The LCSCs were exposed to RNAs and TRAIL followed by staining with dihydroethidium for determining ROS, and propidium iodide (PI) and Annexin V (Thermo Fisher Scientific) were used for determining the extent of apoptosis.
Tumor xenografts nude mice for tumor growth
For the in vivo study, the experimental mice were distributed into four major groups with each group having ten mice. For inducing tumor xenografts, two groups (lentivirus control and TRAIL group) were injected subcutaneously with lentivirus control Huh7 cells, and the other two groups (anti-miR-21-3p and TRAIL-anti-miR-21-3p group) were injected with lentivirus anti-miR-21-3p Huh7 cells. After the tumors were induced, physical parameters such as tumor volume (V) were recorded using calipers. After the tumor volume reached 100 mm3, the animals of TRAIL and TRAIL-anti-miR-21-3p groups received injections of TRAIL (40 µg/kg, intraperitoneal [IP]) once in every 2 days. The volume of tumors was recorded after every 3 days. At the end of the study, ie, after 30 days, the mice were euthanized by the sodium pentobarbitone anesthesia method, which was in accordance to the guidelines for euthanasia as per the Guide for the Care and Use of Laboratory Animals. For finding the number of LCSCs in tumor tissues, the cells obtained from tumor were purified, ie, the tumors were cut and added into collagenase type-3 at a concentration of 1 mg/mL, and about 5 mL was used for each gram of tumor tissue in DMEM containing FBS (10%) for 2 hours at 37°C. The obtained cells were subjected to staining using anti-CD133-FITC stain for 15 minutes, followed by FACS vantage analysis.
Statistical analysis
All the results are expressed as mean ± SD and done in triplicates. The Student’s t-test using GraphPad Prism software was done for statistical analysis. The value of P<0.05 was regarded as significant.
Results
miR-21-3p is upregulated in LCSCs
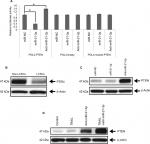
For identifying the involvement of miR-21-3p in LCSCs, qRT-PCR analysis was done in normal and LC cell lines. The results of qRT-PCR suggested higher expression levels of miR-21-3p in selected LC cells compared to normal L-O2 cells (Figure 1A). On studying the expression of miR-21-3p in LCSCs and non-LCSCs of PLC, HepG2 and Huh7, the outcomes suggested (Figure 1B) that miR-21-3p was overexpressed in all the LC cell lines with significant overexpression in Huh7 cells. The results suggested that overexpression of miR-21-3p may be in LCSCs.
Knockdown of miR-21-3p enhances the sensitivity of LCSCs toward TRAIL-induced cell death in vitro
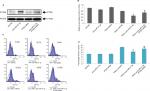
To find the TRAIL-mediated sensitivity of LCSCs and non-LCSCs, the selected Huh7 cell lines were separated as Huh7-LCSCs and non-LCSCs. The two cell lines were exposed to various concentrations of TRAIL. The CD133-Huh7-LCSCs showed a significant reduction in sensitivity compared to non-LCSCs (Figure 2A). For establishing the involvement of miR-21-3p in LCSCs, the non-LCSCs and CD133-Huh7-LCSCs were transfected with miR-21-3p mimics and inhibitors, and the outcomes suggested that transfection of miR-21-3p caused upregulation of miR-21-3p in non-LCSCs and CD133-Huh7-LCSCs, whereas the transfection of anti-miR-21-3p (miR-NC) suppressed the expression of miR-21-3p significantly (Figure 2B) in each cell line. The results of flow cytometry studies suggested that the percentage of LCSCs increased after just a single exposure to TRAIL. We observed that any change in miR-21-3p had no significant effect on the percentage of CD133-Huh7-LCSCs, instead anti-miR-21-3p showed a significant inhibitory role of TRAIL in the CD133-Huh7-LCSCs; on the other hand, the miR-21-3p mimics enhanced the stimulation of CD133-Huh7-LCSCs against TRAIL (Figure 2C and D). The outcomes suggested that the exposure of TRAIL alone resulted in the survival of TRAIL-resistant Huh7-LCSCs and caused death of non-LCSCs sensitive to the treatment of TRAIL. Furthermore, to prove the role of miR-21-3p toward the sensitivity of LCSCs against TRAIL, cell viability assay was done. The LCSCs of the selected three cancer cell lines for the experiment, ie PLC, Huh7 and HepG2 transfected with anti-miR-21-3p, demonstrated significantly greater sensitivity against TRAIL compared to the negative control-treated cells (Figure 2E). We also found that the cells receiving transfection of miR-21-3p mimics exhibited a reversed effect on TRAIL sensitivity against cells treated with negative control. Altogether, outcome of the experiment suggested that suppression of miR-21-3p enhanced the sensitivity of LCSCs against TRAIL-mediated cell death.
Suppression of miR-21-3p enhanced the anti-tumor potential of TRAIL toward LC cell lines (Huh7 CSCs) in vivo
To find the involvement of miR-21-3p inhibitors in cells exposed to TRAIL in vivo, the animal model was created. The nude mice were subcutaneously injected with 5×106 cells of Huh7, which were previously transfected with miR-21-3p or control vector. After grafting cancer cells into mice, the tumors were developed, and the tumor volumes were recorded after every 3 days. The results of tumor volume suggested that the growth of tumors in animals injected with anti-miR-21-3p-transfected Huh7 cells was slow compared to that transfected with control upon both the groups receiving the same treatment of TRAIL (Figure 3A). The results of percentage population of LCSCs in tumor tissues by flow cytometry suggested that increased percentage of cells in tumor tissues of control group compared to tissues of animals injected with anti-miR-21-3p when both groups received the identical treatment of TRAIL (Figure 3B and C). Collectively, the outcomes of the study suggested that suppression of miR-21-3p could enhance the anti-tumor potential of TRAIL and enhance the sensitivity to TRAIL in animal models.
miR-21-3p targets PTEN in Huh7-LCSCs
To interpret the possible role of miR-21-3p in regulating the TRAIL-mediated sensitivity of LCSCs, we utilized TragetScan and PicTar databases. The scans from databases indicated that miR-21-3p targeted the conserved sequence of 3′-UTR of the PTEN-mRNA. The pGL3 luciferase vectors were cloned with PTEN 3′-UTR to affirm that the involvement of miR-21-3p targets directly PTEN-mRNA. The outcomes of dual Luciferase assay demonstrated that co-transfection of luciferase reporter and miR-21-3p mimics with wild-type (wt) of PTEN 3′-UTR caused a significant suppression, whereas the anti-miR-21-3p enhanced the luciferase activities. The empty plasmids do not show any response to miR-21-3p or anti-miR-21-3p (Figure 4A). The outcomes clearly suggest that PTEN as a favorable target of miR-21-3p directly targets the Huh7-LCSCs. Furthermore, we performed protein expression analysis to find the expression levels of PTEN, the results were consistent with Luciferase assay, and due to upregulation of miR-21-3p in Huh7-LCSCs, the level of PTEN was lowered significantly in Huh7-LCSCs compared to that of non-LCSCs (Figure 4B). Furthermore, for more confirmatory evidence about the involvement of miR-21-3p on the level of PTEN, the expression of PTEN was evaluated in Huh7-LCSCs after the cells were transfected with miR-21-3p or anti-miR-21-3p. The blots suggested that anti-miR-21-3p increased the levels, whereas miR-21-3p decreased the expression of PTEN (Figure 4C). We also evaluated the expression of PTEN in tumor tissues of animals, separated as control and TRAIL-treated (lenti-control group), and mice injected with anti-miR-21-3p and anti-miR-21-3p along with the treatment of TRAIL (lenti-anti-miR-21-3p group). The results suggested that the expression of PTEN in miR-21-3p knocked tumor cells and caused a significant increase in the levels of PTEN compared to control (Figure 4D). Altogether, the experiment confirmed that PTEN was a favorable target of miR-21-3p in the LC model in vivo.
Knockdown of miR-21-3p enhances the TRAIL sensitivity of Huh7-LCSCs through the PTEN cascade
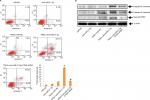
To confirm the involvement of PTEN in cell death mediated by miR-21-3p, using a specific siRNA we suppressed the expression of PTEN (Figure 5A). After transfecting the cells with PTEN siRNA, we found that anti-miR-21-3p caused a significant elevation of TRAIL-mediated toxicity in Huh7-LCSCs, also the expression of anti-miR-21-3p was knocked after transfection with PTEN-siRNA (Figure 5B). We also noted that anti-miR-21-3p caused inhibition in the Huh7-LCSCs population mediated by TRAIL, which was halted by PTEN siRNA (Figure 5C and D). Overall, the outcomes suggested that anti-miR-21-3p enhanced the TRAIL sensitivity of Huh7-LCSCs via enhancing the levels of PTEN.
Suppression of miR-21-3p enhances TRAIL-mediated apoptosis in Huh7-LCSCs
The results have suggested that anti-miR-21-3p do not directly cause apoptosis of HepG2-LCSCs, rather it elevated the effect of TRAIL by causing cell death via apoptosis. Transfection of PTEN siRNA prevented the anti-miR-21-3p-mediated apoptosis of Huh7-LCSCs mediated by anti-miR-21-3p combined with TRAIL, further confirming the results (Figure 6A). We also noticed that suppression of miR-21-3p caused upregulation of caspase-3 and -9, which was induced by the treatment of TRAIL in Huh7-LCSCs; also, the cells transfected with PTEN siRNA resulted in the cleavage of both caspase-3 and -9 mediated by anti-miR-21-3p combined with TRAIL (Figure 6B). Altogether, the findings confirmed that suppression of miR-21-3p in Huh7-LCSCs elevated the apoptosis mediated by TRAIL via the PTEN cascade.
Knockdown of miR-21-3p elevated the TRAIL-mediated apoptosis through the PTEN/PI3K/Akt/Bad cascade
Literature studies have reported that PTEN regulates Akt negatively and also inhibits PI3K.29 We here established the relation between the PTEN/Akt/PI3K cascade and anti-miR-21-3p. In this study, we found that anti-miR-21-3p halted the phosphorylation of Akt and pI3K (Figure 7A), whereas a single exposure of TRAIL failed to affect the expression of Akt or PI3K. As the results depicted that anti-miR-21-3p inhibited phosphorylation of Bad, which according to literature studies is one of the substrates of Akt,30 this may be the mechanism responsible for anti-miR-21-3p-mediated apoptosis of Huh7-LCSCs as described earlier.31 The knockdown of miR-21-3p enhanced the Bad-Bcl-xL and Bad-Bcl-2 heterodimer of Huh7-LCSCs (Figure 7B) and inactivated the Bcl-2 and Bcl-xL. The inactivation of Bcl-2 and Bcl-xl is associated with release of some mitochondria-related apoptotic markers,32 we hence identified Smac/DIABLO, AIF and cytochrome c, and in Huh7-LCSCs we found that expression of all these markers increased significantly (Figure 7C) along with ROS (Figure 7D), after the cells received treatment of anti-miR-21-3p and TRAIL. Altogether, the outcomes suggested that anti-mi-21-3p increases the TRAIL-mediated apoptosis via the PTEN/Akt/PI3K/Bad signaling cascade.
Discussion
A number of literature studies till now have demonstrated the involvement of miRNAs in enhancing the sensitivity of associated treatments in cancer. For instance, miR-122 upon overexpression has been found to reverse the doxorubicin resistance in hepatic cancer via the glucose metabolism pathway.33 Suppression of miR-221 increases the TRAIL-mediated apoptosis in breast cancer by altering the BIM gene.34 Suppression of miR-101 enhances the sensitivity of HCC cell lines via the Mcl-1 pathway.35 Looking into the number of studies suggesting that altering miRNAs could be a vital strategy in countering cancers of lower sensitivity to chemotherapy.
miR-21-3p is the passenger strand and miR-21-5p is the guide strand of miR-21. miR-21 displays oncogenic properties.14 The guide strand ie miR-21-5p is involved with ovarian cancer, breast cancer and lung cancer.20,24,26 We also found an evidence suggesting that both the guide and the passenger strands of miR-21 target the same number of genes and are responsible to regulate expression of their target genes.27 Besides being responsible to regulate equal number of genes, compared to miR-21-5p, ie, guide strand, the role of miR-21-3p, ie, passenger strand, in cancer and resistance studies is not fully elucidated, and also its role in enhancing TRAIL-mediated apoptosis remains unexplored. In this study, we elucidated that miR-21-3p is suppressed in LCSCs. We, in both in vitro and in vivo models, confirmed that suppression of miR-21-3p enhanced the sensitivity toward TRAIL of Huh7-LCSCs. We found that suppression of miR-21-3p halted the consequences of TRAIL by increasing the population of CSCs; subsequently, the absence of miR-21-3p leads to TRAIL-mediated death. The present outcomes confirmed that a miR-21-3p antagonist can be a potential therapeutic approach for countering the character of resistance of LCSCs against TRAIL.
PTEN halts tumors via negatively regulating the PI3K/Akt cascade.36 Studies till now have confirmed that PTEN is deregulated in various cancers, concluding it as one of the potential factors in the process of tumorigenesis.37,38 Previous studies have concluded the role of miRNAs in the regulation of PTEN in cancers, and the PTEN/miRNAs pathway has been identified to be involved in chemosensitivity, growth, invasion and migration.39–41 In this study, we reported that PTEN is suppressed in Huh7-LCSCs and confirmed that miR-21-3p/PTEN is the pathway responsible for sensitivity of CSCs against TRAIL.
BCL2-associated agonist of cell death, also called as Bad, is a member of Bcl-2 family protein and is an Akt substrate; it is a pro-apoptotic marker. Preventing the phosphorylation of Bad leads to apoptosis by inactivating Bcl-xl and Bcl-2. Akt-mediated phosphorylation of Bad leads to the separation of heterodimer of Bcl-2 and Bcl-xL.42 Outcomes of this study demonstrated that the knockdown of miR-21-3p resulted in overexpression of PTEN, and hence halting the Akt/PI3K cascade also keeps Bad in its non-phosphorylated form. From all these events, TRAIL causes mitochondrial apoptosis43 in CSCs guiding the cells to release mitochondria-sourced apoptosis markers such as cytochrome c, ROS, AIF and Smac/DIABLO. Finally, we found that when TRAIL is combined with miR-21-3p, it leads to apoptosis and cleavage of caspase.
Conclusion
This study has produced many evidence to establish that miR-21-3p is associated with TRAIL-mediated apoptosis and also with sensitivity of hepatic LCSCs. Mechanistically, we found that suppression of miR-21-3p leads to TRAIL-mediated apoptosis by halting the PI3K/Bad/Akt cascade involving miR-21-3p/PTEN axis. We conclude that TRAIL along with anti-miR-21-3p could be a new treatment approach for the management of LC.
Abbreviations
CSCs, cancer stem cells; LC, liver cancer; LCSCs, liver cancer stem cells; PTEN, phosphatase and tensin homologue; qRT-PCR, quantitative reverse transcription PCR; TRAIL, TNF-related apoptosis-inducing ligand.
Acknowledgments
We express thanks to the staff and management of The Affiliated Wuxi No.2 People’s Hospital of Nanjing Medical University for providing the necessary facilities. We are also thankful to our funding agencies for providing the essential fund for the project. This study was supported by 1) Gastroenterology of key disciplines from Wuxi (No: DXK002), 2) Academician Workstation of No. 2 People’s Hospital of Wuxi City (No: CYR1705), 3) Wuxi Medical Key Talent Project (No: ZDRC029), 4) Wuxi Sci-Tech Development Fund (No: CSE31N1603) and 5) Technical achievements and appropriate technology promotion project of Wuxi Municipal Commission of Health and Family Planning (No: T201603).
Author contributions
YZ planed the study protocol along with HT, LZ, LG, GW, JN and XT. The experiments were carried out by XT, HT and LZ. LG, GW and JN did the calculation part. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. | ||
Villanueva A, Newell P, Chiang DY, Friedman SL, Llovet JM. Genomics and signaling pathways in hepatocellular carcinoma. Semin Liver Dis. 2007;27(1):55–76. | ||
Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. | ||
Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124(6):1111–1115. | ||
Suetsugu A, Nagaki M, Aoki H, Motohashi T, Kunisada T, Moriwaki H. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Commun. 2006;351(4):820–824. | ||
Ma S, Lee TK, Zheng BJ, Chan KW, Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27(12):1749–1758. | ||
Joshi P, Jeon YJ, Laganà A, et al. MicroRNA-148a reduces tumorigenesis and increases TRAIL-induced apoptosis in NSCLC. Proc Natl Acad Sci U S A. 2015;112(28):8650–8655. | ||
Walczak H, Miller RE, Ariail K, et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med. 1999;5(2):157–163. | ||
Piggott L, Omidvar N, Martí Pérez S, French R, Eberl M, Clarkson RW. Suppression of apoptosis inhibitor c-FLIP selectively eliminates breast cancer stem cell activity in response to the anti-cancer agent, TRAIL. Breast Cancer Res. 2011;13(5):R88. | ||
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. | ||
Gargalionis AN, Basdra EK. Insights in microRNAs biology. Curr Top Med Chem. 2013;13(13):1493–1502. | ||
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. | ||
Ou Y, Zhai D, Wu N, Li X. Downregulation of miR-363 increases drug resistance in cisplatin-treated HepG2 by dysregulating Mcl-1. Gene. 2015;572(1):116–122. | ||
Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37(Pt 4):918–925. | ||
Parker H, Rose-Zerilli MJ, Parker A, et al. 13q deletion anatomy and disease progression in patients with chronic lymphocytic leukemia. Leukemia. 2011;25(3):489–497. | ||
Luo F, Ji J, Liu Y, et al. MicroRNA-21, up-regulated by arsenite, directs the epithelial-mesenchymal transition and enhances the invasive potential of transformed human bronchial epithelial cells by targeting PDCD4. Toxicol Lett. 2015;232(1):301–309. | ||
Yang Q, Xu E, Dai J, et al. miR-21 regulates N-methyl-N-nitro-N’-nitrosoguanidine-induced gastric tumorigenesis by targeting FASLG and BTG2. Toxicol Lett. 2014;228(3):147–156. | ||
Wang G, Wang JJ, Tang HM, To SS. Targeting strategies on miRNA-21 and PDCD4 for glioblastoma. Arch Biochem Biophys. 2015;580:64–74. | ||
Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35(17):5944–5953. | ||
Yang H, Kong W, He L, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68(2):425–433. | ||
Wyman SK, Parkin RK, Mitchell PS, et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS One. 2009;4(4):e5311. | ||
Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. | ||
Integrated Genomic Analysis of Ovarian Carcinoma. Cancer Discov. 2011;1(3):197. | ||
Devulapally R, Sekar TV, Paulmurugan R. Formulation of Anti-miR-21 and 4-Hydroxytamoxifen Co-loaded Biodegradable Polymer Nanoparticles and Their Antiproliferative Effect on Breast Cancer Cells. Mol Pharm. 2015;12(6):2080–2092. | ||
Yang Z, Fang S, Di Y, Ying W, Tan Y, Gu W. Modulation of NF-κB/miR-21/PTEN pathway sensitizes non-small cell lung cancer to cisplatin. PLoS One. 2015;10(3):e0121547. | ||
Pink RC, Samuel P, Massa D, Caley DP, Brooks SA, Carter DR. The passenger strand, miR-21-3p, plays a role in mediating cisplatin resistance in ovarian cancer cells. Gynecol Oncol. 2015;137(1):143–151. | ||
Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35(17):5944–5953. | ||
Lo TF, Tsai WC, Chen ST. MicroRNA-21-3p, a berberine-induced miRNA, directly down-regulates human methionine adenosyltransferases 2A and 2B and inhibits hepatoma cell growth. PLoS One. 2013;8(9):e75628. | ||
Stambolic V, Suzuki A, de la Pompa JL, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95(1):29–39. | ||
Brazil DP, Hemmings BA. Ten years of protein kinase B signalling: a hard Akt to follow. Trends Biochem Sci. 2001;26(11):657–664. | ||
Datta SR, Dudek H, Tao X, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91(2):231–241. | ||
Ye L, Yuan G, Xu F, et al. The small-molecule compound BM-1197 inhibits the antiapoptotic regulators Bcl-2/Bcl-xL and triggers apoptotic cell death in human colorectal cancer cells. Tumour Biol. 2015;36(5):3447–3455. | ||
Pan C, Wang X, Shi K, et al. MiR-122 Reverses the Doxorubicin-Resistance in Hepatocellular Carcinoma Cells through Regulating the Tumor Metabolism. PLoS One. 2016;11(5):e0152090. | ||
Ye Z, Hao R, Cai Y, Wang X, Huang G. Knockdown of miR-221 promotes the cisplatin-inducing apoptosis by targeting the BIM-Bax/Bak axis in breast cancer. Tumour Biol. 2016;37(4):4509–4515. | ||
He H, Tian W, Chen H, Deng Y. MicroRNA-101 sensitizes hepatocellular carcinoma cells to doxorubicin-induced apoptosis via targeting Mcl-1. Mol Med Rep. 2016;13(2):1923–1929. | ||
Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275(5308):1943–1947. | ||
Tay Y, Kats L, Salmena L, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–357. | ||
Xu W, Yang Z, Zhou SF, Lu N. Posttranslational regulation of phosphatase and tensin homolog (PTEN) and its functional impact on cancer behaviors. Drug Des Devel Ther. 2014;8:1745–1751. | ||
Wu W, Yang J, Feng X, et al. MicroRNA-32 (miR-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Mol Cancer. 2013;12:30. | ||
Wang ZX, Lu BB, Wang H, Cheng ZX, Yin YM. MicroRNA-21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting PTEN. Arch Med Res. 2011;42(4):281–290. | ||
Ma F, Zhang J, Zhong L, et al. Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt/β-catenin signaling. Gene. 2014;535(2):191–197. | ||
Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. 1999;13(22):2905–2927. | ||
Lim SC, Parajuli KR, Han SI. The alkyllysophospholipid edelfosine enhances TRAIL-mediated apoptosis in gastric cancer cells through death receptor 5 and the mitochondrial pathway. Tumour Biol. 2016;37(5):6205–6216. | ||
Elshafie HS, Armentano MF, Carmosino M, Bufo SA, De Feo V, Camele I. Cytotoxic Activity of Origanum Vulgare L. on Hepatocellular Carcinoma cell Line HepG2 and Evaluation of its Biological Activity. Molecules. 2017;22(9):E1435. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.