Back to Journals » OncoTargets and Therapy » Volume 12
Supervised Machine Learning Predictive Analytics For Triple-Negative Breast Cancer Death Outcomes
Authors Xu Y, Ju L, Tong J, Zhou C , Yang J
Received 17 July 2019
Accepted for publication 1 October 2019
Published 1 November 2019 Volume 2019:12 Pages 9059—9067
DOI https://doi.org/10.2147/OTT.S223603
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Yucan Xu, Lingsha Ju, Jianhua Tong, Chengmao Zhou, Jianjun Yang
Department of Anesthesiology, First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, People’s Republic of China
Correspondence: Chengmao Zhou; Jianjun Yang
First Affiliated Hospital of Zhengzhou University No. 1 Jianshe East Road, Henan Province, Zhengzhou City 450000, China
Tel +86 187 7675 1816; +86 137 8353 7619
Email [email protected]; [email protected]
Objective: To use machine learning algorithms to predict the death outcomes of patients with triple-negative breast cancer, 5 years after discharge.
Methods: 1570 stage I-III breast cancer patients receiving treatment from Sun Yat-sen Memorial Hospital were analyzed. Machine learning was used to predict the death outcomes of patients with triple-negative breast cancer, 5 years after discharge.
Results: The results showed that platelets, LMR (lymphocyte-to-monocyte ratio), age, PLR (the platelet-to-lymphocyte ratio) and white blood cell counts accounted for a significant weight in the 5-year prognosis of triple-negative breast cancer patients. The results of model prediction indicated that rankings for accuracy among the training group (from high to low) were forest, gbm, and DecisionTree (0.770335, 0.760766, 0.751994, 0.737640 and 0.734450, respectively). For AUC value (high to low), they were forest, Logistic and DecisionTree (0.896673, 0.895408, 0.776836, 0.722799 and 0.702804, respectively). The highest MSE value for DecisionTree was 0.2656, and the lowest MSE value for forest was 0.2297. In the test group, accuracy rankings (from high to low) were DecisionTree, and GradientBoosting (0.748408, 0.738854, 0.738854, 0.732484 and gbm, respectively). For AUC value (high to low), the rankings were GradientBoosting, gbm, and DecisionTree (0.731595, 0.715438, 0.712767, 0.708348 and 0.691960, respectively). The maximum MSE value for gbm was 0.2707, and the minimum MSE value for DecisionTree was 0.2516.
Conclusion: The machine learning algorithm can predict the death outcomes of patients with triple-negative breast cancer 5 years after discharge. This can be used to estimate individual outcomes for patients with triple-negative breast cancer.
Keywords: machine learning, triple-negative breast cancer, 5-year prognosis
Introduction
Breast cancer impairs women’s health worldwide. Among females, it has a global incidence rate higher than any other malignancy. It is a leading cause of death among women aged 40–55, behind only lung, gastric, liver, esophageal and colorectal cancer. Compared with many developed countries in Europe and North America, the age of breast cancer patients in China is trending downward. The epidemiological characteristics of breast cancer in China vary regionally, according to respective ecological, economic and lifestyle differences.
Triple-negative breast cancer (TNBC) refers to breast cancer with negative estrogen receptor, progesterone receptor and human epidermal growth factor receptor 2.1 It accounts for between 10% and 20% of all breast cancers. TNBC has unique biological behaviors and clinicopathological features. Its prognosis is worse than most other cancers. Thus, recurrence-free survival and overall survival are low.2 TNBC patients are resistant to endocrine therapy and molecular-targeted therapy, and therefore most rely on chemotherapy.3
Breast cancer, the most common cancer among women, causes approximately 500,000 deaths per annum. Several effective breast cancer treatments are available, but successful outcomes still depend on early diagnosis and prevention. Therefore, identifying which breast cancer patients remain high-risk after surgery has been the focus of researchers seeking to curtail breast cancer mortality. However, with the advent of the era of big data, professional medical information has been integrated into big data, creating conditions conducive to the integration of machine learning methods in the medical field. Studies4 have shown that correct intelligent recognition can be realized between different subtypes by analyzing breast ultrasound images through machine learning. Furthermore, diagnostic performance is superior to that of standard visual imaging assessments. Moreover, machine learning can identify the mechanisms of new TNBC drugs.5 However to date, no research has used machine learning to study TNBC prognosis. The predictive analysis of breast cancer patient prognosis based on artificial intelligence (machine learning) can also provide reference for clinical patient evaluation, for determining surgical methods, and for developing adjuvant therapies. It can also offer support for the development of treatment plans.
Methods
The original data from Professor Fengxi Su’s articles have been included in BioStudies database. The BioStudies database is a new EMBL‐EBI resource that contains descriptions of biological studies, and links to supporting data in other databases (Including Dryad database). And we have also obtained Professor Fengxi Su’s original data via the following website: https://www.ebi.ac.uk/biostudies/studies/S-EPMC4666347. We conducted a secondary analysis of Professor Fengxi Su’s original data in order to explore the effect of the machine learning algorithms in predicting breast cancer prognosis. We used data from an xlsx file (pone.0143061.s001.xlsx) collected by this website for research and analysis.
Sample
We collected data from 1,570 patients with stage I-III breast cancer who had been treated at Sun Yat-sen Memorial Hospital between January 2000 and December 2010. Five surveillance algorithms were used as a machine learning model to predict the death outcomes of TNBC patients, 5 years after discharge. This study evaluated the predictive effects of the machine learning model on the death outcomes of TNBC patients 5 years of discharge. It thus provides a tool to evaluate TNBC patient prognosis.
Data Source
Our exclusion criteria came from previous studies. 1806 patients with stage I-III breast cancer who had been treated at Sun Yat-sen Memorial Hospital between January 2000 to December 2010 were enrolled. Exclusion and inclusion criteria were as follows. Patients who had received any treatment or had been diagnosed with a metastatic disease prior to surgery or neoadjuvant chemotherapy were excluded. Other exclusion criteria included bilateral breast cancer, male breast cancer, inflammatory breast cancer and long-term cortex hormone therapy. Medical records of the included patients were accessible. A total of 1,570 patients met the inclusion criteria of this study. Whole blood counts and white blood cell count differences for all venous blood samples were tested with a blood analyzer. Estrogen receptor (ER), progesterone receptor (PR) and HER2 analysis were performed by immunohistochemistry (IHC). Tumors with an ER or PR positive that showed a staining intensity greater than or equal to 10% were considered positive. Patients were classified according to the IHC status of their tumors in the following manner: luminal subtype: ER + and/or PR + and HER2-; HER2 positive subtype: HER2 +; and triple negative subtype: ER-, PR- and HER2-. Patient data were obtained through electronic medical records, which had been entered by breast surgeons. To ensure data quality, one researcher was assigned to collect data, and another to check and confirm it.
Overall survival was calculated from the date of pathological diagnosis to death (any cause) or date of the last follow-up. Disease-free survival was calculated from the date of pathological diagnosis to local or distant recurrence, the date of death, or new primary cancer. All patients were followed up every 3 months for the first 3 years, and then followed annually until recurrence or death. The last date of follow-up for all available patients was July 2014.
Abbreviations: Estrogen receptor (ER), progesterone receptor (PR), neutrophil-to-lymphocyte ratio (NLR), lymphocyte-to-monocyte ratio (LMR), platelet-to-lymphocyte ratio (PLR); LightGBM (gbm); Accuracy = all predicted dead samples/total samples; Precision = Predicts death as death/All predictions as death; Recall = Predicts Death as Positive/All Positive True Death; f1_score= Accuracy * recall * 2/(accuracy+recall).
The Machine Learning Algorithm
Logistic regression is a process by which a cost function is established for a regression or classification problem. Then, the optimal model parameters are solved iteratively through the optimization method, and then tested to verify the quality of the solved model.
A decision tree is a predictive model which represents a mapping relationship between object properties and object values. Each node in the tree represents an object; each forked path represents a possible attribute value; and each leaf node corresponds to the object value represented by the path from the root node to the leaf node. A decision tree only has one output. If a complex output is desired, an independent decision tree can be created to handle different outputs. The decision tree is a common technique in data mining, and it can also be used for data analysis and prediction.
Random forests, also known as random decision forests, are collection learning methods that can be used for classification, regression, and other tasks. “Random forest” can be understood literally: the focus is on “random” and “forest”. “Random” indicates random sampling and features, and “forest” refers to a forest of multiple decision trees. A single decision tree may cause over-fitting problems, but these can be addressed with the aggregation results of multiple independent decision-making trees. Therefore, random forest is an integrated learning method.
In the Gradient Boosting framework, the most common base learner is the decision tree. The combination of these two has made the gradient boosted decision tree (GBDT) algorithm famous. GBDTs have 3 features. Firstly, the trees that construct GBDTs are regression decision trees. Secondly, GBDTs use gradient iterative gradient boosting. Thirdly, GBDTs use shrinkage technology.
LigthGBM is a new member of the boosting collection model. It is as efficient as xgboost in implementing GBDT, and in many other aspects, outperforms it. The principles of LigthGBM, GBDT, and xgboot are similar, and they each use the negative gradient of the loss function as an approximate value of the residual of the current decision tree to fit the new decision tree.
Statistical Analysis
The measurement data were expressed in the form of mean standard deviation, and the counting data were expressed in the form of example number (percentage). The data were analyzed with R (version 3.6.0, https://www.r-project.org/) and relevant statistical packages. Obvious outliers were deleted, and the missing variable values were filled with multiple imputation. The train_test_split function of the sklearn package in Python (Python Software Foundation, version 3.6) was used to randomly group the data into a training set and a verification set at an 8:2 ratio. This was used to repeat the training of the model to verify its stability. Scikit Learning (https://github.com/scikit-learn/scikit-learn)] was used for machine learning. By using Python 3.6, model parameter range was preset based on previous application experience, and the model classification results were reported with the classification_report function. The matplotlibAPI provided support for mapping to draw the ROC curve predicted by each model. The overall classification effect of each model was evaluated by the area under the curve (AUC), and the classification effect of the cut points selected by the model was described with accuracy and F1-score.6 Mean square error refers (MSE) to the expected value of the square of the difference between the estimated value of the parameter and the true value of the parameter. MSE can evaluate the degree of change in the data. The smaller the MSE value, the more accurate the prediction model was at describing the experimental data. AUC values were interpreted as: 0.5–0.7: low effect; 0.7–0.85: general effect; 0.85–0.95: good effect; 0.95–1: the effect was very good.
Background Information
The sample contained 1,143 patients with triple-negative breast cancer who had survived 5 years after discharge, and 425 patients who died 5 years after discharge. There was a statistical difference in PLATELET between the two groups, with a P-value of 0.038. The two groups’ average tumor size also showed a significant statistical difference, p < 0.001. There was also a significant statistical difference in NSAID medication between the two groups, P < 0.001. The NLR, PLR and LMR values of the two groups had no statistical difference, P-value > 0.05 (See in Table 1).
 |
Table 1 Baseline Data |
The death outcome for triple-negative breast cancer 5 years after discharge was the dependent variable (0 = no, 1 = yes), and all 19 factors were independent variables for LightGBM algorithm analysis. The results showed that platelets, LMR, age, PLR and white blood cell counts influenced 5-year prognosis and death of triple-negative breast cancer patients. Figure 1 presents the positive correlation between age, NSAID, lymphocyte count and 5-year prognosis and death for breast cancer. Chemotherapy and PR are each negatively correlated with 5-year prognosis and death. (See in Figures 1 and 2)
 |
Figure 1 Correlation analysis of various factors. |
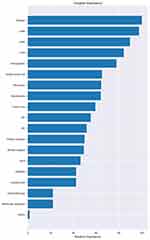 |
Figure 2 Variable importance of features included in the machine learning algorithm for predicting triple-negative breast cancer five-year mortality. |
A flow chart of data processing by machine learning is shown in Appendix Figure 1.
The final parameters through which the prediction results of the machine learning model passed were: Logistic regression-penalty=‘l2ʹ, tol=0.0001, C=0.7, fit_intercept=True, intercept_scaling=1, class_weight=None, max_iter=100, multi_class=“ovr”, verbose=0, warm_start=False,n_jobs=−1; GradientBoosting-learning_rate=0.01, n_estimators= 100, min_samples_split=10, min_samples_leaf=1, subsample=0.5, max_depth=5; gbm-boosting_type= “gbdt”, reg_alpha= 0.001, reg_lambda= 0.8, learning_rate=0.1, max_depth=1, n_estimators=100, and objective=“binary” (See Appendix Table 1).
The model’s prediction results show that in the training group, accuracy, from highest to lowest, went forest, gbm, gradientboost, Logistic and DecisionTree (distribution: 0.770335, 0.760766, 0.751994, 0.737640 and 0.734450). AUC value from highest to lowest was forest, gbm, gradientboost, Logistic and DecisionTree (distribution: 0.896673, 0.895408, 0.776836, 0.722799 and 0.702804). The highest MSE value for DecisionTree was 0.2656, and the lowest MSE value for forest was 0.2297. Forest had the highest precision value (1) and DecisionTree had the lowest precision value (0.518325). The highest recall value for DecisionTree was 0.291176, and the lowest recall value for gbm was 0.123529. The highest f1_score value for DecisionTree was 0.372881, and the lowest f1_score value for gbm was 0.218750 (See Table 2 and Figure 3).
 |
Table 2 Forecast Results For Training Group |
In the test group, accuracy, from highest to lowest, went DecisionTree, forest, Logistic, gradientboost and gbm (distribution: 0.748408, 0.738854, 0.738854, 0.732484 and 0.729299). AUC value from highest to lowest was gradientboost, Logistic, forest, gbm and DecisionTree (distribution: 0.731595, 0.715438, 0.712767, 0.708348 and 0.691960). The maximum MSE value for gbm was 0.2707, and the minimum MSE value for DecisionTree was 0.2516. Forest’s precison value was 1 at its highest and gbm’s precision value was 0.500000 at its lowest. The highest recall value for DecisionTree was 0.294118, and the lowest recall value for gbm was 0.023529. The highest f1_score value for DecisionTree was 0.387597, and the lowest f1_score value for gbm was 0.044944 (See Table 3 and Figure 4).
 |
Table 3 Forecast Results For Testing Group |
Discussion
The continuous clinical trials of TNBC and the publication of research results have resulted in several breakthroughs in the systemic treatment of advanced TNBC.7,8 Moreover, with advancing genetic testing technology, the genotyping of TNBC has also been clear, but the efficient therapy is still absent.9 TNBC patients have bleak prognosis—the 5-year survival rate is below 15%, and the time to recurrence is short. Death peaks 5 years after diagnosis. The incidence of brain metastases in patients is high, so patients may die of rapid metastasis.10 Therefore, a valid evaluation method is important for TNBC prognosis.
In our study, considering the accuracy, precision and AUC of the training group and the test group, the forest algorithm came in first in overall performance; the logistic algorithm was the most stable. Judging by AUC value only, forest and gbm performed best in the training group. In the training group and the test group, the AUC values of Logistic, forest, gradient boost and gbm algorithms were all above 0.7, which can predict the death outcome of triple-negative breast cancer 5 years after discharge. At the same time, the results of the LightGBM algorithm show that platelets, LMR, age, PLR and white blood cell counts influence the death outcome of triple-negative breast cancer 5 years after discharge; age, NSAID and lymphocyte count are positively correlated with the death outcome of triple-negative breast cancer 5 years after discharge; chemotherapy and PR are negatively correlated with the death outcome of triple-negative breast cancer 5 years after discharge.
Lymphocytes are involved in immune response. The stronger the local immunological effect, the less favorable the tumor growth and metastasis. Conversely, the stronger the local immunosuppressive effect, the more favorable the tumor invasion and metastasis.11 Tumor-infiltrating lymphocytes (TIL) are important immune cells in tumors, and low lymphocyte counts are thought to be responsible for the lack of immunological response that leads to low survival rates for various cancers.12,13 Higher peripheral lymphocyte counts predict lower mortality in TNBC that may be cured early. This suggests that immune function can enhance early TNBC treatment.14 This is similar to the results in our study.
As part of the inflammatory response, thrombocytosis is common in cancer patients. Platelets in solid tumors promote tumor cell growth by secreting various angiogenic and tumor growth factors. In addition, platelets protect cancer cells from natural killer-mediated cleavage, and promote distant metastasis by activating the Smad and NF-κB pathways.15 The meta-analysis reported by Wei S et al16 suggests that preoperative PLR might be an independent risk factor in pancreatic cancer prognosis. A meta-analysis by Zhang et al17 including 19 articles and 6,314 esophageal cancer patients concluded that PLR might be an important prediction biomarker in esophageal cancer patients. Additionally, LMR is a poor predictor of luminal A and HER-2 overexpressed TNBC with molecular subtype distribution different from PLR.18 Our results reached the same conclusion.
Studies have shown that regular NSAID use improves colorectal cancer survival rates.19 However, studies have shown that the use of NSAID is associated with increased endometrial cancer-specific mortality, especially in patients with type I tumors.20 In our study, we found a positive correlation between the use of NSAID and the death outcome of TNBC patients 5 years after discharge.
The hematopoiesis age of patients at the onset of TNBC determines the aggressiveness and progression of the tumor.21 TNBC in elderly patients is a subtype of invasive breast cancer, but chemotherapy intervention is necessary regardless of age.22 Our results show a negative correlation between chemotherapy and death outcomes among TNBC patients 5 years after discharge.
Nomograms are a traditional statistical method for evaluating breast cancer prognosis. After a review of previous studies23–27 in which nomograms were used to predict breast cancer prognosis, we found that the nomogram prediction established by Wen et al performed the best, with a value of 0.789. The random forest algorithm in our study had a maximum value of 0.896673. Moreover, the advantage of machine learning algorithms lies in their capacity to automatically process large samples of data, learn automatically, and optimize algorithms based on past experience, thus performing better on the ensuing prediction.
With the explosive growth of medical data, machine learning has gained 5 distinct advantages over traditional statistical analysis. (1) Precision; Machine learning uses data to identify an optimized decision engine to solve problems. With the increase in data, precision is improved. (2) Automation; Since the results can only be effective and abandoned, machine learning automatically learns new patterns. This means that users can embed machine learning directly into automatic workflows. (3) Speed; Machine learning produces results within milliseconds upon data entry, allowing the system to react in real time; many data-driven problems can be solved with machine learning. (4) Wide selectivity; Machine learning models are built from their own data and can be optimized with any evaluation criteria. (5) Large-scale processing capacity; As the business continues to evolve, machine learning can manage data growth issues. Some machine learning algorithms are able to process large amounts of data with cloud computing. Moreover, many recent studies28,29 have indicated that machine learning can predict early biochemical recurrence after robot-assisted prostatectomy. Additionally, a prognostic genome for patients with epithelial ovarian cancer has been developed through a machine learning model.
This study has both advantages and disadvantages. Machine learning modeling is a new technology that has not been widely used in TNBC prognosis. However, machine learning algorithms have been applied to complex situations with successful results. For example, machine learning models based on texture features extracted by wavelet transform can improve the ability to distinguish TNBC from benign fibrous adenomas.30 Our results suggest that this technology has the potential to improve TNBC prognosis in the future. Nonetheless, this is a hypothesis generation study, and external validation of the model is critical to validating its utility. The limitation of this study is its retrospective data collection, and as such, our research may be biased because of incomplete data. Also, this study only focused on 5-year prognosis, and did not address longer-term prognosis.
Conclusion
In summary, platelets, LMR, age, PLR, and white blood cell counts may be associated with death outcomes in TNBC patients 5 years after discharge. Machine learning modeling techniques can produce better predictive models for 5-year death outcomes in TNBC patients. These techniques may help in estimating individual outcomes and selecting better therapies for TNBC patients.
Ethics Approval And Consent To Participate
The ethics approval for the original study by Professor Fengxi Su came from Sun Yat-sen Memorial Hospital.
Consent For Publication
All consent of the personal data was obtained from the corresponding person.
Availability Of Data And Material
The datasets used and/or analysed for the present study are available from the corresponding author upon reasonable request.
Acknowledgements
Professor Fengxi Su’s previously published articles have shown that preoperative NLR elevation is an independent prognostic biomarker of disease-free survival and overall survival in TNBC. At the same time, in order to find, reuse and quote the research data freely, the data from this retrospective analysis article is also included in the BioStudies database31 (https://www.ebi.ac.uk/biostudies/studies/S-EPMC4666347). We are grateful to Professor Fengxi Su for sharing his data32 and allowing us to use them for research.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wang Y, Zhang T, Kwiatkowski N, et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell. 2015;163(1):174–186. doi:10.1016/j.cell.2015.08.063
2. Barbie TU, Alexe G, Aref AR, et al. Targeting an IKBKE cytokine network impairs triple-negative breast cancer growth. J Clin Invest. 2014;124(12):5411–5423. doi:10.1172/JCI75661
3. OShaughnessy J, Osborne C, Pippen JE, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. New Eng J Med. 2011;364(3):205–214. doi:10.1056/NEJMoa1011418
4. Wu T, Sultan LR, Tian J, et al. Machine learning for diagnostic ultrasound of triple-negative breast cancer. Breast Cancer Res Treat. 2019;173:365–373. doi:10.1007/s10549-018-4984-7
5. Athreya AP, Gaglio AJ, Cairns J, et al. Machine learning helps identify new drug mechanisms in triple-negative breast cancer. IEEE Trans Nanobiosci. 2018;17:251–259. doi:10.1109/TNB.2018.2851997
6. Powers DM. Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. Mach Learn Technol. 2011;2(1):37–63.
7. O’Sullivan H, Collins D, O’Reilly S. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2019;380:986.
8. Denkert C, Liedtke C, Tutt A, et al. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet. 2017;389:2430–2442. doi:10.1016/S0140-6736(16)32454-0
9. Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750. doi:10.1172/JCI45014
10. Abdulkarim BS, Cuartero J, Hanson J, et al. Increased risk of locoregional recurrence for women with T1-2N0 triple-negative breast cancer treated with modified radical mastectomy without adjuvant radiation therapy compared with breast-conserving therapy. J Clin Oncol. 2011;29(21):2852–2858. doi:10.1200/JCO.2010.33.4714
11. Pauken KE, Wherry EJ. Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 2015;36(4):265–276. doi:10.1016/j.it.2015.02.008
12. Hoffmann TK, Dworacki G, Tsukihiro T, et al. Spontaneous apoptosis of circulating T lymphocytes in patients with head and neck cancer and its clinical importance. Clin Cancer Res. 2002;8(8):2553–2562.
13. Vayrynen JP, Tuomisto A, Klintrup K, et al. Detailed analysis of inflammatory cell infiltration in colorectal cancer. Br J Cancer. 2013;109(7):1839–1847. doi:10.1038/bjc.2013.508
14. Afghahi A, Purington N, Han SS, et al. Higher absolute lymphocyte counts predict lower mortality from early-stage triple-negative breast cancer. Clin Cancer Res. 2018;24:2851–2858. doi:10.1158/1078-0432.CCR-17-1323
15. Schlesinger M. Role of platelets and platelet receptors in cancer metastasis. J Hematol Oncol. 2018;11:125. doi:10.1186/s13045-018-0669-2
16. Song W, Tian C, Wang K, et al. Preoperative platelet lymphocyte ratio as independent predictors of prognosis in pancreatic cancer: a systematic review and meta-analysis. PLoS One. 2017;12(6):e0178762. doi:10.1371/journal.pone.0178762
17. Zhang X, Wang Y, Zhao L, et al. Prognostic value of platelet-to-lymphocyte ratio in oncologic outcomes of esophageal cancer: a systematic review and meta-analysis. Int J Biol Markers. 2018;33:335–344.
18. Zenan H, Zixiong L, Zhicheng Y, et al. Clinical prognostic evaluation of immunocytes in different molecular subtypes of breast cancer. J Cell Physiol. 2019;234:20584–20602. doi:10.1002/jcp.v234.11
19. Coghill AE, Newcomb PA, Campbell PT, et al. Prediagnostic non-steroidal anti-inflammatory drug use and survival after diagnosis of colorectal cancer. Gut. 2011;60:491–498. doi:10.1136/gut.2010.221143
20. Brasky TM, Felix AS, Cohn DE, et al. Nonsteroidal anti-inflammatory drugs and endometrial carcinoma mortality and recurrence. J Natl Cancer Inst. 2017;109:1–10.
21. Marsh T, Wong I, Sceneay J, et al. Hematopoietic age at onset of triple-negative breast cancer dictates disease aggressiveness and progression. Cancer Res. 2016;76:2932–2943. doi:10.1158/0008-5472.CAN-15-3332
22. Königsberg R, Pfeiler G, Klement T, et al. Tumor characteristics and recurrence patterns in triple negative breast cancer: a comparison between younger (<65) and elderly (≥65) patients. Eur J Cancer. 2012;48:2962–2968. doi:10.1016/j.ejca.2012.04.019
23. Wen J, Yang Y, Liu P, et al. Development and validation of a nomogram for predicting survival on the base of modified lymph node ratio in breast cancer patients. Breast. 2017;33:14–22. doi:10.1016/j.breast.2017.01.017
24. Shunrong L, Zhao J, Zhu L, et al. Development and validation of a nomogram predicting the overall survival of stage IV breast cancer patients. Cancer Med. 2017;6:2586–2594. doi:10.1002/cam4.1224
25. Lai J, Pan Z, Chen P, et al. Development and validation of a nomogram incorporating axillary lymph node ratio to predict survival in node-positive breast cancer patients after neoadjuvant chemotherapy. Jpn J Clin Oncol. 2019;49:22–28. doi:10.1093/jjco/hyy181
26. Li X, Huang H, Lin Q, et al. Validation of a breast cancer nomogram to predict lymphedema in a Chinese population. J Surg Res. 2017;210:132–138. doi:10.1016/j.jss.2016.11.009
27. Xiong Z, Deng G, Huang X, et al. Score for the survival probability in metastasis breast cancer: a nomogram-based risk assessment model. Cancer Res Treat. 2018;50:1260–1269. doi:10.4143/crt.2017.443
28. Wong NC, Lam C, Patterson L, et al. Use of machine learning to predict early biochemical recurrence after robot-assisted prostatectomy. BJU Int. 2019;123:51–57. doi:10.1111/bju.14477
29. Lu T-P, Kuo K-T, Chen C-H, et al. Developing a prognostic gene panel of epithelial ovarian cancer patients by a machine learning model. Cancers (Basel). 2019;11(2). doi:10.3390/cancers11020270.
30. Moon WK, Huang Y-S, Lo C-M, et al. Computer-aided diagnosis for distinguishing between triple-negative breast cancer and fibroadenomas based on ultrasound texture features. Med Phys. 2015;42:3024–3035. doi:10.1118/1.4921123
31. Sarkans U, Gostev M, Athar A, et al. The BioStudies database-one stop shop for all data supporting a life sciences study. Nucleic Acids Res. 2018;46:D1266–D1270. doi:10.1093/nar/gkx965
32. Jia W, Wu J, Jia H, et al. The peripheral blood neutrophil-to-lymphocyte ratio is superior to the lymphocyte-to-monocyte ratio for predicting the long-term survival of triple-negative breast cancer patients. PLoS One. 2015;10:e0143061. doi:10.1371/journal.pone.0143061
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


