Back to Journals » International Journal of Nanomedicine » Volume 14
Superparamagnetic iron oxide nanoparticle-mediated expression of miR-326 inhibits human endometrial carcinoma stem cell growth
Authors Gao Y, Qian H, Tang X, Du X, Wang G, Zhang H, Ye F, Liu T
Received 5 January 2019
Accepted for publication 20 March 2019
Published 18 April 2019 Volume 2019:14 Pages 2719—2731
DOI https://doi.org/10.2147/IJN.S200480
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Mian Wang
Yongtao Gao,1 Haiyang Qian,2 Xue Tang,1 Xiling Du,3 Gang Wang,2 Hairong Zhang,2 Fei Ye,2 Te Liu4
1The International Peace Maternity and Child Health Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, 200030, People’s Republic of China; 2Department of Imaging, Dahua Hospital, Shanghai, 200237, People’s Republic of China; 3School of Life Science and Technology, Tongji University, Shanghai 200092, People’s Republic of China; 4Shanghai Geriatric Institute of Chinese Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai 200031, People’s Republic of China
Background: Previously, our group confirmed the presence of a subset of cancer stem cells in the tissues of endometrial carcinoma (ie, human endometrial carcinoma stem cells [HuECSCs]). However, the mechanisms by which microRNAs regulate the growth of HuECSCs remain elusive.
Methods: We loaded miR-326 onto superparamagnetic iron oxide nanoparticles (miR-326@SPION) and transfected them into HuECSCs.
Results: In the present study, we found that the expression levels of members of the G-protein coupled receptor 91 (GPR91)/signal transducer and activator of transcription 3 (STAT3)/vascular endothelial growth factor (VEGF) pathway were significantly elevated in CD44+/CD133+ HuECSCs. Luciferase reporter assays indicated that the succinate receptor 1 (SUCNR1) gene, also known as the G-protein coupled receptor 91 (GPR91) gene, was one of the potential targets of miR-326. Transmission electron microscopy revealed that the SPIONs could cross the cell membrane and accumulate in the cytoplasm. The overexpression of miR-326 significantly inhibited the proliferation and cell cycle progression of HuECSCs in vitro. MiR-326 overexpression also effectively inhibited the invasion and angiogenic capacities of HuECSCs in the extracellular matrix. Meanwhile, miR-326 overexpression significantly inhibited the tumorigenicity and tumour neovascularization capacity of HuECSCs in nude mice. Both quantitative real-time PCR and Western blotting confirmed that overexpression of miR-326 significantly reduced the expression of members of the GPR91/STAT3/VEGF pathway in HuECSCs, and the activity (level of phosphorylation) of key molecules in this pathway was also reduced.
Conclusion: Collectively, we confirmed that SPIONs are highly efficient nanocarriers for nucleic acids, on which the loading of miR-326 inhibited the activation of the GPR91/STAT3/VEGF signaling pathway and significantly attenuated the activity of stem cells in endometrial carcinoma, both in vitro and in vivo.
Keywords: human endometrial carcinoma stem cells, superparamagnetic iron oxide nanoparticles, microRNA, succinate receptor 1
Introduction
Endometrial carcinoma, a common type of gynecological cancer, is a relatively indolent tumour that is usually difficult to detect at an early stage. Meanwhile, type II endometrial carcinoma is often characterized by negative expression of estrogen receptors, a high degree of malignancy, poor differentiation, and unsatisfactory treatment outcome with chemotherapy, which pose severe threats to women’s physical health.1–4 Previously, we reported that in the type II endometrial carcinoma cell lines KLE and ANCA3, the CD44+/CD133+ cell subset exhibited potential cancer stem cell properties, including rapid self-proliferation, invasiveness, drug resistance, and high tumorigenicity.1–4 We also confirmed that the differential expression of the DLK1-DIO3 Genomic Imprinted MicroRNA Cluster is closely related to the degree of malignancy of CD44+/CD133+ HuECSCs.2,5 Thus, some tumour inhibit-microRNAs could suppress the proliferation and invasion of HuECSCs, which have a significant negative regulatory effect. And these microRNAs are potential and novel targets for HuECSCs treatment drug development.
Magnetic nanomaterials have particle sizes between 0 and 100 nm and are composed of iron, cobalt, nickel, and their alloys, and can produce magnetism directly or indirectly.6–8 Among this array of magnetic nanoparticles, iron oxide nanoparticles have been widely studied because of their high saturation magnetization values, low toxicity, readily available raw materials, and high surface reactivity.6–8 In recent years, the use of iron oxide nanoparticles as gene carriers has received increasing attention. Magnetic nanoparticles with sizes less than 20 nm often exhibit superparamagnetism. Superparamagnetic iron oxide nanoparticles (SPIONs) are currently a hot topic in gene carrier research because they possess controllable features, and are stable and easily modified.6–8 Upon binding to plasmid DNA and short interfering RNA (siRNA), SPIONs can deliver the bound nucleic acids into mammalian cells under the influence of an external magnetic field. The internal and external barriers of cells are overcome via magnetic adsorption, which helps to increase local DNA concentrations and improve the transfection efficiency.6–8 Studies have demonstrated that modifying the SPION surface with cationic liposomes or cationic polymers, such as polyethyleneimine, dendrimers, glucose, and chitosan, further enhances the interactions between the nanomaterials and nucleic acids to be transfected, which improves the transfection efficiency.6–8 Previously, we confirmed that SPIONs bind effectively to microRNAs or siRNAs to mediate their expression in cells, in which they either inhibit tumour cell proliferation and invasion, or promote stem cell differentiation efficiency.6–8
According to above evidences, we speculated that some microRNAs could negatively regulate the growth of endometrial cancer stem cells. If SPIONs were used as a carrier, could it realize the loading and transporting of tumour suppressor microRNAs in endometrial cancer stem cells? In the present study, we used SPIONs to mediate the efficient expression of microRNA-326 (miR-326@SPION) in CD44+/CD133+ HuECSCs and found that they inhibited the growth of HuECSCs by suppressing the expression of members of the G-protein coupled receptor 91 (GPR91)/signal transducer and activator of transcription 3 (STAT3)/vascular endothelial growth factor (VEGF) pathway.
Materials and methods
Isolating CD44+/CD133+ HuECSCs
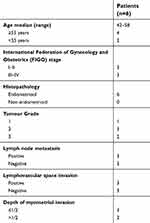
Endometrial carcinoma tissues were collected from the International Peace Maternity and Child Health Hospital (Shanghai, China) between May 2017 and Dec 2017 (Table 1). CD44+/CD133+ cell subpopulations were isolated from tumour samples as previously described.2,3 The study involving human tissues has been obtained according to consent regulation and approved by the Ethics Review Committee of Shanghai Geriatric Institute of Chinese Medicine of Research in Human Production authorized by Shanghai Municipal Government; meanwhile, the informed consent has been provided by all patients conducted in accordance with the Declaration of Helsinki. And, we have received written informed consent from patients. After isolation, single cells were plated at 1000 cells/mL in Dulbecco’s modified Eagle’s medium (DMEM): F12 (HyClone) supplemented with 10 ng/mL basic fibroblast growth factor, 10 ng/mL epidermal growth factor, 5 μg/mL insulin, and 0.5% bovine serum albumin (all Sigma-Aldrich, St. Louis, MO, USA). CD44+/CD133+ cells were cultured under the above conditions as non-adherent spherical clusters (HuECSCs). CD44-/CD133- cells were cultured under general conditions as adherent clusters (HuECCs). All cells were cultured under the same conditions until passage 4 before further experiments.
 | Table 1 Characteristics of the patients’ cohort |
Bioinformatic analysis
Sequence alignment, using bioinformatics analysis software (TargetScan Release 7.2 - Prediction of microRNA targets tools,
Luciferase reporter assay
All cells were seeded at 30,000 cells/well in 24-well cell culture plates. Using Lipofectamine 2000 Reagent, 400 ng of pcDNA-miR-326, pcDNA-miR-mut, or pcDNA, and 20 ng of pGL6-GPR91-3UTR, or pGL6-GPR91-mut, or pGL6 was transfected into each group of cells, separately. At 48 h after transfection, the luciferase activity in each group of cells was examined using a dual-luciferase reporter assay system (Promega). All plasmid DNA was puchased from NOVOBIO (NOVOBIO Biotechnology Co., Ltd., Shanghai, China).
SPION- induced microRNA transfection into cells
SPIONs were purchased from NOVOBIO (NOVOBIO Biotechnology Co., Ltd., Shanghai, China). Transfection was performed according to the manufacturer’s protocol and as previously published. Five microliters of 0.2 mM SPIONs and 5 μL of 10 μM pcDNA-miR-326 plasmid DNA or pcDNA-miR-mut plasmid DNA (NOVOBIO) were thoroughly mixed, vortexed for 10 s, and allowed to stand at room temperature for 20 min. Then, 10 μL of pcDNA-miR-326@SPIONs or pcDNA-miR-mut@SPIONs, prepared in the previous step, were combined with 90 μL of serum-free DMEM:F12 (1:1), added to the cells (1×104 cells/mL, and incubated for 72 h at 37 °C in an atmosphere of 5% CO2.
Cell proliferation assay
Cells were seeded at 2×103 per well in 96-well plates and cultured in DMEM supplemented with 10% fetal bovine serum (FBS) at 37 °C under 5% CO2 until 85% confluence. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma Chemicals) (5 mg/mL) was added at different time points and incubated for a further 4 h. The reaction was terminated by adding 150 μL/well dimethyl sulfoxide (Sigma Chemicals). Cells were lysed for 15 min, and the plates were gently shaken for 5 min. Absorbance at 490 nm was determined using a Model 680 Microplate Reader (Bio-Rad).
RNA extraction and reverse transcription
RNA extraction was performed according to the manufacturer’s protocol of the RNAprep pure Tissue Kit (TIANGEN Biotech (Beijing) Co., Ltd. China). Briefly, approximately 20 mg of human tissue samples was obtained and mixed with 800 μL of lysate for homogenization. The supernatants were collected and added to 200 μL of chloroform. The samples were mixed by inversion and then subjected to centrifugation at 13,400× g for 15 min at 4 °C, after which the supernatants were collected. Absolute ethanol at 2 times the volume of supernatants was added. After mixing by inversion, the mixture was centrifuged at 13,400× g for 30 min at 4 °C. The RNA pellets were resuspended in 500 μL of 75% ethanol and then centrifuged at 13,400× g for 5 min at 4 °C. The excess liquid was removed, and the pellets were completely dissolved in 300 μL of DEPC water. One microliter of RNA solution was used to measure the OD260/OD280 values (generally maintained between 1.8 and 2.0) and to verify the purity and total concentration of the RNA. Reverse transcription was performed according to the manufacturer’s protocol of the miRcute miRNA First-strand cDNA kit (TIANGEN Biotech). Briefly, 20 μL of total RNA (100 ng/μL) was mixed with 25 μL of 2× miRNA RT Reaction Buffer, 4 μL of 1× miRNA RT Enzyme Mix, and 6 μL of RNase-free deionized water. The following reaction was performed in a PCR machine: 42 °C for 60 min for poly(A) tailing and reverse transcription of miRNA; followed by 95 °C for 3 min for enzyme inactivation.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was performed according to the manufacturer’s protocol of the miRcute miRNA qPCR Detection kit (TIANGEN Biotech). Briefly, the reaction comprised: 10 μL of 2× miRcute Plus miRNA Premix (with SYBR), 1×1 μL (10 μM) of forward primer, 1×1 μL (10 μM) of reverse primer, 4 μL of first strand cDNA from miRNA, and 4 μL of deionized water. The following reactions were carried out in a real-time PCR machine: 40 cycles of 95 °C for 15 min, 94 °C for 20 s, and 60 °C for 34 s. The fluorescence values were recorded. The sequences of qPCR primers were as follows: miR-326-F: CCUCUGGGCCCUUCCUCCAG; miR-326-R: GCTGTCAACGATACGCTACCTA; Gpr91-F: TCAAGGGATCAAGTCTTCCAA; Gpr91-R: CCAGCCAGTTTTTGCAAGTT; Vegf-F: CTACCTCCACCATGCCAAGT; Vegf-R: AGCTGCGCTGATAGACATCC; Kdr-F: CCTGTATGGAGGAGGAGGAA; Kdr-R: CGGCTCTTTCGCTTACTGTT; Stat3-F: CCTCTGCCGGAGAAACAG; Stat3-R: CTGCTCCAGGTACCGTGTGT; Mapk14-F: TTCTCCGAGGTCTAAAGTA; Mapk14-R: TGCCGAGCCAGTCCAAAA; 18s rRNA-F: CAGCCACCCGAGATTGAGCA; 18s rRNA-R: TAGTAGCGACGGGCGGTGTG.
Western blotting
Western blotting was performed as previously described.2,9,10 Briefly, total protein from each group of cells was subjected to SDS-PAGE on a 12% denaturing gel. Afterwards, the proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore). After blocking and washing the membrane, primary antibodies9 were added and the membrane was incubated at 37 °C for 15 min. After extensive washing, secondary antibodies were added and incubated at 37 °C for 45 min. The membrane was subjected to four 14 min washes with Tris-buffered saline-Tween 20 (TBST) at room temperature. The membrane was then developed using an enhanced chemiluminescence (ECL) kit (Pierce Biotechnology) and exposed to X-ray film (Sigma-Aldrich Chemical) for visualization. The gray levels of western blotting protein band were quantified by using ImageJ software (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, MD, USA). To determine relative expression levels of target proteins, the formula was used as follows: (Experiment group_target protein gray level value/Experiment group_GAPDH protein gray level)/(Control group_target protein gray level value/Control group_GAPDH protein gray level) value. The protein levels were calibrated based on levels of GAPDH. The information of antibodies were as follows: Rabbit anti-p38 antibody [E229] (ab170099), Mouse anti-p38MAPK (phospho T180+ Y182) antibody [M139] (ab45381), Rabbit anti-VEGF Receptor 2 antibody (ab2349), Rabbit anti-GPR91 antibody (ab41505), Mouse anti-STAT3 (phospho Y705) antibody [EP2147Y] (ab76315), Rabbit anti-STAT3 antibody [EPR787Y] (ab68153), Mouse anti-VEGF 165A antibody [6B7] (ab69479), Rabbit anti-GAPDH antibody [EPR16891] (ab181602), Goat anti-Rabbit IgG H&L (HRP) (ab97051), Mouse anti-Ki67 antibody [B126.1] (ab8191), Goat anti-Mouse IgG H&L (HRP) (ab6789), Goat anti-Rabbit IgG H&L (Alexa Fluor® 488) (ab150077), Goat Anti-Mouse IgG H&L (Cy3) preadsorbed (ab97035). All antibodies were purchased from Abcam.
Propidium iodide staining and flow cytometry
Propidium iodide (PI) staining and flow cytometry were performed as previously described.2 Briefly, 5×105 cells/ml were harvested and fixed in 1 mL of ice-cold 70% ethanol for 48 h. The cells were centrifuged at 1500 r/min for 5 min at 4 °C. The cell pellets were collected, treated with PI staining solution (Sigma Chemicals), and incubated in the dark at 4 °C for 30 min. A flow cytometer (BD FACSAria) was used to determine the cell cycle distribution of each group of cells, and data analyses were performed using the CellQuest software.2
Hematoxylin and eosin staining
Tissue samples were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. The paraffin-embedded tissues were cut into 4-μm sections using a microtome, and the sections were affixed onto glass slides. Subsequently, the sections were dewaxed using xylene and subjected to dehydration in an ethanol gradient. The sections were stained with hematoxylin (H) for 5 min at room temperature, and then 1% ethanol was added for 30 s for differentiation. Afterwards, aqueous ammonia was added for 1 min for blueing, followed by rinsing in distilled water for 5 min. Subsequently, the sections were stained with eosin (E) for 2 min at room temperature and then rinsed with distilled water for 2 min. Then, decolorization over an ethanol gradient was performed, and xylene was added for 2 min for clearing. Finally, the sections were sealed and mounted with neutral resin.
Immunofluorescence staining
Briefly, fresh tissues were immersed in 4% paraformaldehyde (Sigma-Aldrich) for fixation at room temperature for 30 min. The tissues were then dehydrated in an ethanol gradient, embedded in paraffin, sectioned (thickness: 6 μm), and immersed in xylene for dewaxing. Tissue sections were blocked with immunohistochemical blocking solution (Beyotime Biotechnology Co., Ltd., Zhejiang, China) at 37 °C for 30 min. The blocking solution was then discarded, and the sections were washed three times at room temperature for 5 min each with immunohistochemical washing solution (Beyotime Biotechnology). Then, primary antibodies were added and incubated at 37 °C for 45 min. After incubation, the antibody solution was discarded, and the sections were washed three times at room temperature for 5 min each with immunohistochemical washing solution (Beyotime Biotechnology). Then, secondary antibodies were added and the tissues were incubated at 37 °C for 45 min. After incubation, the antibody solution was discarded, and the sections were washed three times at room temperature for 5 min each with immunohistochemical washing solution (Beyotime Biotechnology). Finally, immunofluorescence blocking solution (Sigma-Aldrich) was added, and the sections were mounted.
Northern blotting
Northern blotting was performed as previously described.2,10 Briefly, total RNA was extracted from all groups of cells using a Trizol kit. Following quantification, 20 μg of high-quality total RNA was subjected to gel electrophoresis on a 7.5 M urea-12% formaldehyde (PAA) denaturing gel. Afterwards, the RNA was transferred to a Hybond N+ nylon membrane (Amersham, Freiburg, Germany). The membrane was cross-linked under 1200 mjoule/cm2 of UV for 30 s. An antisense DNA probe against miR-326 was used for hybridization to detect the expression status of miR-326 (5’-GGAGACCCGGGAAGGAGGTC-3’). After hybridization and washing, the membrane was exposed to Kodak XAR-5 films (Sigma-Aldrich Chemical) for 20–40 h. As a positive control, the human U6 snRNA probe (5′-GCAGGGGCCATGCTAATCTTCTCTGTATCG-3′) was used for hybridization in all membranes. The exposure time for the U6 snRNA probe was maintained between 15 and 30 min.2,10 The gray levels of Northern blotting hybridization bands were quantified by using ImageJ software. To determine relative levels of hybridisation signal, the formula was used as follows: Target hybridisation signal gray value/U6 snRNA gray value. The hybridisation signal levels were calibrated based on levels of U6 snRNA.
Transwell assay
Briefly, 200 μL of serum-free cell culture medium containing 2000 cells/mL were seeded into the upper chambers of Transwell plates with pore sizes of 8.0 μm. Then, 600 μL of complete medium containing 10% FBS was added to the lower chambers of the Transwell plates. The cells were cultured at 37 °C for 48 h in an atmosphere of 5% CO2. The cells adhering to the membrane surface were fixed with 4% paraformaldehyde at room temperature for 30 min and were then stained with 4,6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich Chemical) for 10 min. The total number of cells was calculated by counting the number of cells in three non-overlapping fields under a microscope.
In vivo xenograft experiments
In vivo xenograft experiments were performed as previously described.2 Briefly, after transfection with plasmids, 1×105 cells/mL from each group at logarithmic growth phase were harvested and inoculated subcutaneously into BALB/Cnu/nu mice. Each group comprised six mice (6-8-week-old female BALB/Cnu/nu mice were provided by the Experimental Animal Centre of Fudan University). After 10 weeks of monitoring, the mice were sacrificed, and the tumours were removed. The tumours were weighed, and the volumes were calculated using the following formula: Tumour volume (mm3) = (ab2)/2 (a: the longest axis (mm), b: the shortest axis (mm)). All the animal experiments were conducted in accordance with the guidelines of the NIH for the care and use of laboratory animals. The study protocol was also approved by the Committee on the Use of Live Animals in Teaching and Research, Shanghai Geriatric Institute of Chinese Medicine, Shanghai, China.2
Statistical analysis
Each experiment was performed as least three times; data are presented as mean ± the standard error (SE) where applicable. Differences were evaluated using Student’s t-tests. P-values <0.05 were considered statistically significant.
Results
GPR91 is a specific target of miR-326
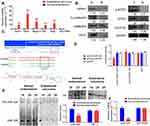
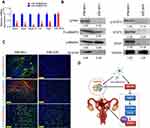
The GPR91/STAT3/VEGF signaling pathway plays an important role in tumour proliferation; therefore, we evaluated the expression of key molecules in this signaling pathway in normal endometrium and endometrial carcinoma tissues. The results from qPCR and western blotting demonstrated that the mRNA and protein expression levels of GPR91, STAT3, mitogen-activated protein kinase P38 alpha (p38MAPK; also known as mitogen-activated protein kinase 14 (MAPK14)), vascular endothelial growth factor receptor 2 (VEGFR2, also known as kinase insert domain receptor (KDR)), and VEGF were significantly higher in endometrial carcinoma tissues than in the normal endometrium (Figure 1A and B). Bioinformatic analysis showed perfect complementary pairing between the mature miR-326 and seven bases of a specific site on GPR91 (SUCNR1) (+121 bp to +127 bp), suggesting that GPR91 could be one of the targets of miR-326 (Figure 1C). In addition, comparisons of the genomes revealed that the target of miR-326 was highly conserved, and the sequence was “CCCAGAG” in both rodents and mammals (Figure 1C). Subsequently, luciferase reporter assays showed that the overexpression of wild-type (WT) miR-326 in cells significantly reduced the activity of luciferase in the construct carrying WT GPR91, while the luciferase activity in the remaining combinations was not affected (Figure 1D). Northern blotting results showed that the hybridization signal of miR-326 was significantly lower in endometrial carcinoma tissues compared with that in normal tissues (Figure 1E). Meanwhile, qPCR results also showed that expression level of miR-326 was significantly lower in endometrial carcinoma tissues than in normal tissues (Figure 1F). These results indicated that the GPR91/STAT3/VEGF signaling pathway is significantly activated in endometrial carcinoma tissues; in addition, GPR91 is a specific target of miR-326.
SPIONs can accumulate in HuECSCs
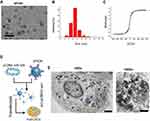
Analyses showed that the size of the SPION core was between 2 and 14 nm, and its hysteresis loop exhibited a symmetrical distribution (Figure 2A–C). When the SPION surface was modified with polyethyleneimine (PEI), it could bind efficiently with the miR-326-expressing plasmid to form a “nucleic acid-nanoparticle” complex (Figure 2D). When the SPION surface was loaded with large quantities of plasmid DNA, the hydrodynamic size of the SPIONs increased significantly (48.0 nm before loading; 161.5 nm after loading). Plasmid DNA is negatively charged; therefore, the zeta potential was also reduced significantly after adsorption (+30.5 mV before loading; +26.3 mV after loading). Nonetheless, a relatively strong positive charge was maintained; thus, the nanoparticles still displayed good stability and no coagulation occurred. CD44+/CD133+ HuECSCs were isolated from primary endometrial carcinoma cells via flow cytometry and cultured in suspension (Figure 2E). Then, SPIONs were added to the culture medium of the HuECSCs. Transmission electron microscopy revealed that large quantities of dense electron clouds were aggregated in microvesicles within the cytoplasm of HuECSCs, which were presumed to be SPIONs (Figure 2E). These data demonstrated that SPIONs could accumulate in HuECSCs.
mir-326@SPION significantly inhibits the proliferation, invasion, and angiogenesis of HuECSCs
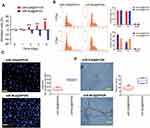
We examined the effects of miR-326@SPION on the growth of HuECSCs in vitro. The results from the MTT assays showed that the rate of inhibition of HuECSCs proliferation gradually increased in the miR-326@SPION group 2 days after SPION transfection and was significantly higher than that in the miR-mut@SPION group (control group). In addition, the inhibition of cell proliferation was significantly time-dependent (Figure 3A). The results of flow cytometry showed that at 3 days after SPION transfection, the proportion of HuECSCs in the G0/G1 phase was significantly higher in the miR-326@SPION group than that in the control group, whereas the proportion of cells in the S phase was significantly lower than that in the control group (Figure 3B). This suggested that the cell cycle of HuECSCs in the miR-326@SPION group was significantly blocked in the G0/G1 phase. Meanwhile, the Transwell chamber cell invasion assay showed that the number of migrated HuECSCs was significantly lower in the miR-326@SPION group than in the control group at 3 days after SPION transfection (Figure 3C). Furthermore, when human umbilical vein endothelial cells (HUVECs) were transfected with miR-326@SPION, their ability to migrate and form capillaries in the extracellular matrix were significantly reduced compared with that of cells in the miR-mut@SPION transfection group (Figure 3D). These results showed that miR-326@SPION significantly inhibits the proliferation, invasion, and angiogenesis of HuECSCs in vitro.
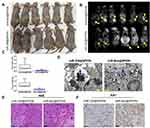
Subsequently, we inoculated both groups of cells (miR-326@SPION and miR-mut@SPION) onto the back of nude mice. The nude mice were sacrificed at around week 10. All the nude mice that were inoculated with miR-mut@SPION-HuECSCs developed relatively large tumours on their backs. The nude mice inoculated with miR-326@SPION-HuECSCs also developed tumours on their backs; however, the tumours were significantly smaller in terms of both size and weight compared with those in the control group (Figure 4A and C). Magnetic resonance imaging (MRI) scans performed on living nude mice revealed significant high-intensity signals at the tumour sites on the backs of the mice, suggesting the presence of metallic materials (Figure 4B). Subsequently, the tumour tissues were isolated. Transmission electron microscopy revealed the presence of high-density electron clouds resulting from the aggregation of circular particles in the xenograft tumours derived from both cell groups, suggesting the presence of metallic nanoparticles (Figure 4D). Lastly, pathological examination of the tumour sections via H&E staining showed that the tumours derived from both groups of cells displayed the pathological features of type II endometrial carcinoma (Figure 4E). Immunohistochemical staining results indicated that the percentage of Ki67-positive cells in tumours derived from miR-326@SPION-HuECSCs was significantly lower than in those derived from miR-mut@SPION-HuECSCs (Figure 4F). These results indicated that miR-326@SPION significantly inhibits the tumorigenicity of HuECSCs in nude mice.
mir-326@SPION significantly inhibits the expression of the GPR91/STAT3/VEGF signaling pathway
The results of qPCR showed that the mRNA expression levels of GPR91, STAT3, p38MAPK (MAPK14), VEGFR2 (KDR), and VEGF were significantly lower in HuECSCs transfected with miR-326@SPION compared with those in the control group (Figure 5A). Meanwhile, western blotting analysis also showed that the protein levels of GPR91, STAT3, p38MAPK, VEGFR2, and VEGF were significantly lower in HuECSCs transfected with miR-326@SPION compared with those in the control group (Figure 5B). Meanwhile, the levels of the phosphorylated products of STAT3, and p38MAPK, ie, p-STAT3, and p-p38MAPK were also significantly lower compared with those in the HuECSCs transfected with miR-mut@SPION (Figure 5B). Furthermore, immunofluorescence staining was performed on the xenograft tumour tissues of nude mice. The results showed that expression of proteins of the GPR91/STAT3/VEGF signaling pathway was significantly lower in tumours derived from miR-326@SPION-HuECSCs compared with that derived from miR-mut@SPION-HuECSCs (Figure 5C). In addition, the expression of VEGFR2 was also significantly reduced, suggesting that miR-326@SPION significantly inhibited angiogenesis in endometrial carcinoma tissues. These results suggested that miR-326@SPION could significantly inhibit the expression of members of the GPR91/STAT3/VEGF signaling pathway in HuECSCs.
Discussion
Previously, studies from our group confirmed the presence of a subset of cancer stem cells in endometrial carcinoma tissues, and preliminarily identified certain microRNAs that may affect the growth and tumorigenicity of this group of cells.1–3,10 However, the growth of tumour cells is substantially different from that of normal cells, particularly in terms of energy metabolism and angiogenesis.1 Tumour cells are often in a highly active state of metabolism and angiogenesis. Therefore, blocking these abnormal biological behaviors may significantly inhibit tumour growth. During our investigation of the processes of vascular ageing and formation, we noted that intervention with oxidized low-density lipoprotein in apolipoprotein E (ApoE)−/- mice could inhibit the expression of the GPR91/STAT3/VEGF pathway through the activation of miR-758, which promoted the ageing of vascular endothelial cells and inhibited angiogenesis.9 The results of that study provided us with new insights into the targets and pathways for suppressing endometrial carcinoma stem cells. GPR91 is also known as succinate receptor 1. GPR91 is expressed in various tissues and organs, such as the retina, heart, liver, kidney, white adipose tissue, and dendritic cells derived from human monocytes.11–17 Succinate is a direct ligand for GPR91.17–19 It is also an intermediate metabolite of the Krebs cycle and is essential for the synthesis of adenosine triphosphate in mitochondria and for several metabolic pathways.11–17 In addition to its key role in energy metabolism, succinate can also serve as a signaling molecule through binding to and activating its specific receptor.11–17 The activation of GPR91 induced by local extracellular succinate can inhibit the degradation of white adipose tissue, mediate retinal angiogenesis, increase renin release in the glomerular endothelium, promote the secretion of proinflammatory cytokines from dendritic cells to activate T helper cells, and regulate liver fibrosis. Mu et al found that succinate upregulates the expression of vascular endothelial growth factor through a hypoxia inducible factor 1 subunit alpha (HIF-1α)-independent mechanism, which in turn activates STAT3 and extracellular regulated kinase 1/2.12 Agiar et al reported that succinate may cause cardiac hypertrophy in a CuSnR1-dependent manner, and that serum levels of succinate are significantly elevated in patients with cardiac hypertrophy associated with acute and chronic ischemic diseases.15 In addition, the activation of GPR91 triggers the phosphorylation of extracellular regulated kinase 1/2, the expression of calcium/calmodulin-dependent protein kinase IIδ, and the cytoplasmic translocation of histone deacetylase 5, all of which are signaling events associated with hypertrophy.11–17 In cases of limited blood supply, such as that in ischemia, succinate can be produced through an alternate route other than the Krebs cycle, and the blood concentration of succinate may increase.11–17 Taken together, it has been demonstrated that succinate-mediated activation of the GPR91/STAT3/VEGF signaling pathway has great significance in the development of pathological damage in cardiovascular diseases. However, there is no report on its effects on the regulation of tumour cells.1–3,10 On the present study, we identified a microRNA, miR-326, which could bind to the specific site in the 3′ UTR of GPR91 mRNA, leading to the targeted silencing of GPR91 expression. Through various molecular biology techniques, we showed that miR-326 was expressed at low levels in endometrial carcinoma tissues but at high levels in normal tissues, suggesting that this microRNA was likely to be a tumour suppressor. We also found that the expression pattern of the GPR91/STAT3/VEGF signaling pathway was opposite to that of miR-326, suggesting that the microRNA could potentially have a negative regulatory effect on this pathway. We chose a special nanoparticle, SPION, to verify our hypothesis. Our previous studies demonstrated that SPIONs could effectively deliver siRNAs or microRNAs into cells to achieve the efficient expression of these molecules.6–8 Therefore, we constructed the miR-326@SPION conjugate and transfected it to endometrial carcinoma stem cells. In vitro and in vivo experiments showed that the overexpression of miR-326 significantly attenuated the proliferation, division, invasion, and tumorigenicity of HuECSCs. At the same time, the number of nascent blood vessels in the tumours developed in the nude mice was also markedly reduced. This series of experiments confirmed that miR-326@SPIONs could significantly inhibit the growth of HuECSCs in vitro and in vivo. Next, we further analyzed the underlying mechanism. Through the use of molecular biology techniques, we found that the overexpression of miR-326 significantly reduced the expression of members of the GPR91/STAT3/VEGF pathway in HuECSCs, and concurrently reduced the activity (level of phosphorylation) of key molecules in this pathway. Based on the results from previous studies, activation of the GPR91/STAT3/VEGF pathway always has positive effects on angiogenesis and cell proliferation. Our study indicated that when miR-326 significantly inhibited the expression of GPR91 and the activation of the GPR91/STAT3/VEGF pathway, it could also reduce the activity of cancer stem cells. Therefore, miR-326 and GPR91 are potential and important targets for the regulation of endometrial carcinoma (Figure 5D).6–8
In summary, our study elucidates two key points. First, GPR91 is a potential target of miR-326, and GPR91/STAT3/VEGF is its direct downstream pathway. Second, overexpression of miR-326 could significantly attenuate the activity of endometrial carcinoma stem cells by inhibiting the activation of the GPR91/STAT3/VEGF signaling pathway.
Acknowledgments
This work was supported by grants from the Medical and Industrial Interdisciplinary of Shanghai Jiao Tong University Foundation (YG2014MS33), the Shanghai Natural Science Foundation (16ZR1434000), and from the projects sponsored by the Development Fund for Shanghai Talents (2017054).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387(10023):1094–1108. doi:10.1016/S0140-6736(15)00130-0
2. Gao Y, Liu T, Huang Y. MicroRNA-134 suppresses endometrial cancer stem cells by targeting POGLUT1 and Notch pathway proteins. FEBS letters. 2015;589(2):207–214. doi:10.1016/j.febslet.2014.12.002
3. Gao Y, Liu T, Cheng W, Wang H. Isolation and characterization of proliferative, migratory and multidrug-resistant endometrial carcinoma-initiating cells from human type II endometrial carcinoma cell lines. Oncol Rep. 2012;28(2):527–532. doi:10.3892/or.2012.1807
4. Dowdy SC, Glaser GE. Adjuvant therapy for women with high-risk endometrial carcinoma. Lancet Oncol. 2018;19(3):268–269. doi:10.1016/S1470-2045(18)30091-3
5. Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13(6):e249–e258. doi:10.1016/S1470-2045(12)70073-6
6. Pan Z, Shi Z, Wei H, et al. Magnetofection based on superparamagnetic iron oxide nanoparticles weakens glioma stem cell proliferation and invasion by mediating high expression of microRNA-374a. J Cancer. 2016;7(11):1487–1496. doi:10.7150/jca.15515
7. Liu T, Zhang H, Zheng J, et al. SPION-mediated miR-141 promotes the differentiation of HuAESCs into dopaminergic neuron-like cells via suppressing lncRNA-HOTAIR. J Cell Mol Med. 2018;22(4):2299–2310. doi:10.1111/jcmm.13512
8. Fang K, Liu P, Dong S, et al. Magnetofection based on superparamagnetic iron oxide nanoparticle-mediated low lncRNA HOTAIR expression decreases the proliferation and invasion of glioma stem cells. Int J Oncol. 2016;49(2):509–518. doi:10.3892/ijo.2016.3571
9. Zhang H, Zheng J, Lin J, et al. miR-758 mediates oxLDL-dependent vascular endothelial cell damage by suppressing the succinate receptor SUCNR1. Gene. 2018;663:1–8. doi:10.1016/j.gene.2018.04.029
10. Cheng W, Liu T, Wan X, Gao Y, Wang H. MicroRNA-199a targets CD44 to suppress the tumorigenicity and multidrug resistance of ovarian cancer-initiating cells. FEBS J. 2012;279(11):2047–2059. doi:10.1111/j.1742-4658.2012.08589.x
11. Vargas SL, Toma I, Kang JJ, Meer EJ, Peti-Peterdi J. Activation of the succinate receptor GPR91 in macula densa cells causes renin release. J Am Soc Nephrol. 2009;20(5):1002–1011. doi:10.1681/ASN.2008070740
12. Mu X, Zhao T, Xu C, et al. Oncometabolite succinate promotes angiogenesis by upregulating VEGF expression through GPR91-mediated STAT3 and ERK activation. Oncotarget. 2017;8(8):13174–13185. doi:10.18632/oncotarget.14485
13. Littlewood-Evans A, Sarret S, Apfel V, et al. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J Exp Med. 2016;213(9):1655–1662. doi:10.1084/jem.20160061
14. He W, Miao FJ, Lin DC, et al. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature. 2004;429(6988):188–193. doi:10.1038/nature02488
15. Favret S, Binet F, Lapalme E, et al. Deficiency in the metabolite receptor SUCNR1 (GPR91) leads to outer retinal lesions. Aging (Albany NY). 2013;5(6):427–444. doi:10.18632/aging.100563
16. de Castro Fonseca M, Aguiar CJ, Da Rocha Franco JA, Gingold RN, Leite MF. GPR91: expanding the frontiers of Krebs cycle intermediates. Cell Commun Signal. 2016;14:3. doi:10.1186/s12964-016-0126-1
17. Aguiar CJ, Rocha-Franco JA, Sousa PA, et al. Succinate causes pathological cardiomyocyte hypertrophy through GPR91 activation. Cell Commun Signal. 2014;12:78. doi:10.1186/1478-811X-12-2
18. Tannahill GM, Curtis AM, Adamik J, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496(7444):238–242. doi:10.1038/nature11986
19. Mills E, O‘Neill LA. Succinate: a metabolic signal in inflammation. Trends Cell Biol. 2014;24(5):313–320. doi:10.1016/j.tcb.2013.11.008
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.