Back to Archived Journals » Open Access Insect Physiology » Volume 5
Superoxide dismutase activities in the midgut of Helicoverpa armigera larvae: identification and biochemical properties of a manganese superoxide dismutase
Authors Lomate P, Sangole K, Sunkar R, Hivrale V
Received 5 March 2015
Accepted for publication 10 June 2015
Published 4 August 2015 Volume 2015:5 Pages 13—20
DOI https://doi.org/10.2147/OAIP.S84053
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Guy Smagghe
Purushottam R Lomate,1,2,* Kirti P Sangole,1,* Ramanjulu Sunkar,3 Vandana K Hivrale1
1Department of Biochemistry, Dr Babasaheb Ambedkar Marathwada University, Aurangabad, Maharashtra, India; 2Department of Entomology, Iowa State University, Ames, IA, 3Department of Biochemistry and Molecular Biology, Oklahoma State University, Stillwater, OK, USA
*These authors contributed equally to this work
Abstract: Insects possess a complex network of enzymatic antioxidant systems protecting against reactive oxygen species generated during stress. Superoxide dismutases (SODs) are vital antioxidant enzymes converting superoxide into oxygen and hydrogen peroxide. Helicoverpa armigera is a polyphagous insect and has developed resistance to various phytochemicals and pesticides. Although SODs are studied in several insect species, their characterization has been reported in only a few insect species. Here, we attempted to identify and characterize SOD activity from H. armigera. Electrophoretic separation followed by nitroblue tetrazolium staining revealed the presence of five SOD activity isoforms in the midgut of H. armigera larvae. Total SOD activity was measured at the different larval developmental stages and found maximum at fourth instar and further declined at sixth instar. Total SOD activity was significantly increased by metal ions Mn, Fe, Zn, and Cu and reduced by Ca and Pb. The isoform with highest activity was identified as manganese SOD (MnSOD) because it was found insensitive to H2O2. The identified MnSOD was partially purified and characterized. After gel filtration chromatography, specific activity of the purified enzyme was found to be 6,348.0 U/mg/min with 26.61% yield. The molecular mass of purified enzyme was calculated to be ~29.3 kDa by sodium dodecyl sulfate polyacrylamide gel electrophoresis. The optimum pH and temperature for the SOD activity were found to be around 11 and 60°C, respectively. The study could be useful to further understand the functional significance of SODs in the antioxidant mechanism of H. armigera.
Keywords: antioxidant mechanism, H. armigera, superoxide dismutases
Introduction
Induction of excessive reactive oxygen species (ROS) in insects is attributed to their exposure to various stress factors throughout the lifetime. Insects have evolved complex network of enzymatic antioxidant systems to protect against ROS.1 Superoxide dismutases (SODs) are a major component of the antioxidant enzyme system of insects, which convert superoxide into oxygen and hydrogen peroxide.2–4
Helicoverpa armigera (Lepidoptera: Noctuidae) (Hübner) is the most polyphagous, widely distributed, and destructive insect foraging on many important crop plants and causes heavy economic losses.5,6 It is continuously exposing to variety of phytotoxins and insecticides commonly used for pest control. These insecticides increase the level of free radicals and influence antioxidant defense mechanism in cells.7 Insecticides are also known to cause oxidative stress by inducing lipid peroxidation and protein oxidation, and consequently induce production of ROS.8–10 Plant prooxidants such as quercetin (a flavonoid) or xanthotoxin (a furanocoumarin) and harmine (a β-carboline alkaloid) are known to produce toxic ROS in the insect gut.11–13 A key role of SOD in protecting insects from prooxidant toxicity was evident when its inhibition resulted in the enhanced toxicity toward prooxidants.12 Altogether, SODs are principle components of insect antioxidant system, which hold a strong potential for study. Moreover, the polyphagous insects including H. armigera which are continuously being exposed to different phytochemicals and chemical pesticides must possess strong antioxidant mechanisms, where SODs play a significant role. Therefore, in this study, we focused to characterize the SOD proteins from H. armigera to understand the biochemistry of these important enzymes.
Several common forms of SOD exist whose active site uses copper and zinc, or manganese, iron, or nickel. Thus, there are three major families of SODs, depending on the protein fold and the metal cofactor: the Cu/Zn type (which binds both copper and zinc), Fe and Mn types (which bind either iron or manganese), and the Ni type (which binds nickel). Biochemical characterization of SODs along with the identification of individual enzyme isoform is an interesting aspect of research in insects. Although several studies from various insect species have reported the molecular characterization and responses of SODs against different types of stresses,14–18 knowledge on the identification and characterization of SODs in H. armigera is lacking. Similarly, the role of SODs in insect resistance mechanism against insecticides is also not completely understood yet. There are few studies showing elevated activities and expression of SOD genes in insects after feeding on insecticides. For example, a recent microarray analysis revealed the elevated transcription of SODs in pyrethroid-resistant strains of Anopheles arabiensis.19 The transcript levels of an SOD were found to be higher in the pesticide-resistant strains of Cimex lectularius, suggesting that these proteins are potentially involved in the elimination of intracellular toxins, which eventually contributes to the pesticide resistance.20 Studies on antioxidant enzymes in H. armigera are potentially essential while understanding its resistance mechanism. The present investigation reports the characterization of SOD activities and identification of a manganese SOD (MnSOD) from H. armigera.
Materials and methods
Chemicals
Following chemicals were obtained from Sigma-Aldrich (St Louis, MO, USA): nitroblue tetrazolium (NBT), xanthine oxidase, xanthine, ethylenediaminetetraacetic acid (EDTA), Coomassie Brilliant Blue R-250, sodium dodecyl sulfate (SDS), riboflavin, bovine serum albumin, hydrogen peroxide, and Sephadex G-50. Medium-range (14.3–97.4 kDa) protein molecular weight markers were obtained from Merck-Genei (Bangalore, India). All other chemicals used were of high analytical grade.
Insects
H. armigera larvae were obtained from the International Crop Research Institute for Semi Arid Tropics (Hyderabad, India) and reared on artificial diet (12 hours light:12 hours dark). The diet was prepared according to the methods described previously.21–23
Preparation of midgut extract
A total of 100 actively feeding fourth instar larvae were immobilized by keeping at -20°C for 30 minutes and dissected. Midguts were removed, weighed, and homogenized with a prechilled mortar and pestle in 1:6 (w/v) volumes of ice-cold 0.1 M Tris–HCl buffer (pH 8).22–24 The homogenate was centrifuged at 9,168× g for 20 minutes at 4°C. The supernatant was collected and divided into small aliquots and stored at -20°C until use. Protein concentration from the supernatant was estimated by the method of Lowry using bovine serum albumin as standard.25
Visualization of H. armigera SOD activity
Native slab polyacrylamide gel electrophoresis (PAGE) was used to separate SODs.26 Visualization of separated SOD activity isoforms was performed using previously described method.27 Briefly, midgut extract (40 μg proteins) was loaded onto the gel, and electrophoresis was carried out at the constant current of 20 mA. After electrophoresis, the gel was washed with distilled water (D/W) and equilibrated in 0.1 M phosphate buffer (pH 8) for 10 minutes. The buffer was removed, and gel was soaked in 1.23 mM NBT solution for 15 minutes in light. Excess NBT was removed by washing the gel with D/W, and then, the gel was soaked in 0.1 M Tris–HCl buffer (pH 8) containing 2.8×10−2 mM riboflavin and 28 mM tetramethylethylenediamine (TEMED) for 15 minutes in dark. The gel was thoroughly rinsed with D/W to remove excess TEMED and riboflavin, and then, it was further transferred into D/W and developed under light (photochemical reaction) until activity bands were observed.
Total SOD activity during developmental stages of H. armigera larvae
Total SOD activity was evaluated from third, fourth, fifth, and sixth instar larvae by using electrophoretic analysis and enzyme assays. SOD activity assay was performed by the method of Imanari et al.28 The reaction mixture was prepared with 2.4 mL of 0.1 M glycine–NaOH buffer (pH 11.0), and 0.1 mL each of 3 mM xanthine, 3 mM EDTA, 0.75 mM NBT, and 0.1 mL of enzyme extract. Reaction was initiated by the addition of xanthine oxidase (56 mU/mL) and continued at 37°C for 20 minutes. Then, reaction was terminated by using 0.84 mM allopurinol, and the change in absorbance was measured at 560 nm. Visualization of SOD activity isoforms was performed as described in the previous section.
Effect of metal ions on total SOD activity
Effect of metal ions (Mn2+, Ca2+, Zn2+, Fe2+, Cu2+, and Pb2+) on total SOD activity was investigated using spectrophotometric assay. Metal ions in their respective concentrations (5 mM each) were pre-incubated with enzyme in a reaction mixture, and the assay was carried out as described earlier for SOD activity assay.
Characterization of SOD activity isoforms
SOD activity isoforms showing activity in the presence of 5 mM H2O2 are considered as MnSODs.29,30 H. armigera midgut extract was mixed with 5 mM H2O2, and the mixture was incubated at 37°C for 30 minutes and loaded onto 10% polyacrylamide gel. Visualization of SOD activity isoforms was carried out as described previously. The SOD isoform b with major activity was characterized as MnSOD, since it was not inhibited by H2O2.
Partial purification of H. armigera MnSOD
H. armigera midgut (20 g) was homogenized with a prechilled mortar and pestle in 1:6 (w/v) volumes of ice-cold 0.1 M Tris–HCl buffer (pH 7.8). Homogenate was centrifuged at 9,168× g for 20 minutes at 4°C, and supernatant was collected. To the supernatant, 1:2 (v/v) volumes of chilled acetone was added and kept for overnight at –20°C. The mixture was then centrifuged at 9,168× g for 20 minutes, acetone was removed by air drying, and precipitate was collected. The precipitate was dissolved in minimum amount of 0.1 M Tris–HCl buffer (pH 7.8) and centrifuged at 9,168× g for 20 minutes, and supernatant was collected. This supernatant (5 mL) was loaded onto a Sephadex G-50 column (1.2 cm ×50 cm) equilibrated with 0.1 M Tris–HCl buffer (pH 7.8). Column was eluted at 4°C with a flow rate of 0.5 mL/min with 0.1 M Tris–HCl buffer (pH 7.8), and fractions of 2 mL were collected. Elution of proteins was monitored at 280 nm. All collected fractions were screened for MnSOD activity.
Molecular mass determination
Molecular mass of purified MnSOD was determined by SDS-PAGE according to the procedure of Laemmli.31 Purified protein (10 μg) was loaded onto 12% SDS polyacrylamide gel with standard molecular weight markers (14.3–97.4 kDa), and electrophoresis was carried out at a constant current of 20 mA. After electrophoresis, the gel was stained with Coomassie Brilliant Blue R-250 and distained to visualize protein bands.
Optimum temperature and pH of MnSOD activity
The stability of MnSOD at different temperatures (10°C–90°C) was evaluated. Purified enzyme was incubated at various temperatures ranging from 10°C to 90°C with 10°C intervals. The reaction mixture was prepared with 2.4 mL of 0.1 M glycine–NaOH buffer (pH 11.0), and 0.1 mL each of 3 mM xanthine, 3 mM EDTA, 0.75 mM NBT, and 0.1 mL of preheated enzyme. Reaction was initiated by the addition of xanthine oxidase (56 mU/mL) and continued at 37°C for 20 minutes. Then, reaction was terminated by using 0.84 mM allopurinol, and the change in absorbance was measured at 560 nm. The reaction rate was linear with time, protein content, and substrate concentration. The activity of MnSOD in various buffers was also evaluated by pre-incubating the reaction mixture with 200 μL of each buffer (0.1 M, pH 2.0–12.0 buffers: i) glycine–HCl, pH 2.0 and 3.0; ii) acetate buffer, pH 4.0 and 5.0; iii) phosphate buffer, pH 6.0 and 7.0; iv) Tris–HCl, pH 8.0 and 9.0; and v) glycine–NaOH, pH 10.0, 11.0, and 12.0). The assay was carried out as described above for optimum temperature.
Statistical analysis
All the experiments were carried out at least three times with three biological replicates using 30 larvae for each replicate. One-way analysis of variance and Student’s t-tests were performed to verify the significance of the observed differences in enzyme activities during different instars and with various metal ions using SPSS, version 15·0 (SPSS Inc., Chicago, IL, USA).
Results
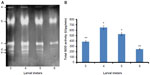
Larval stages of H. armigera and total SOD activity
Five SOD activity isoforms were detected in H. armigera midgut (Figure 1A), which were numbered as a–e. SOD b and c showed higher activity as compared to SOD a, d, and e. Nevertheless, SOD b showed maximum activity, while isoform a displayed minimum activity as compared to the remaining isoforms (Figure 1A). Qualitative and quantitative estimation of total SOD activity was executed at different developmental stages (third to sixth instar) of H. armigera larvae. Results revealed that total SOD activity was directly proportional to the growth of larvae up to fourth instar and found to be 600 U at this stage (t-test, P<0.05). However, the activity was further slightly declined at fifth instar (500 U) and observed to be lowest at sixth instar (200 U) of the larval development (t-test, P<0.05) (Figure 1B). Similar results were evident from electrophoretic detection and enzyme assays (Figure 1).
Effect of metal ions on total SOD activity
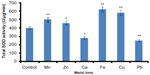
Effect of metal ions on H. armigera SOD activity was evaluated using the larval gut extract. Protein extraction was done with actively feeding fourth instar H. armigera larvae by expecting the maximum enzyme activities at this stage of growth. The method of extraction was thoroughly described in the “Materials and methods” section. Total SOD activity was significantly increased in the presence of metal ions such as Fe, Cu, Mn, and Zn, whereas the metal ions Ca and Pb were found to be responsible for decrease in total SOD activity, suggesting that these metals are not good cofactors for SOD activity (Figure 2). The highest total SOD activity was recorded with Fe, whereas minimum enzyme activity was recorded in the presence of Pb as compared to the activity without metal ions (Figure 2; t-test, P<0.05).
Identification of H. armigera MnSOD
Usually, inhibition assay with H2O2 is used as a marker to characterize the SODs. SOD which is found insensitive in the presence of higher concentration of H2O2 (5 mM) is considered as MnSOD. In the present study, among the five SOD activity isoforms detected, only SOD b was not inhibited, while all the remaining isoforms were found to be inhibited in the presence of H2O2 (Figure 3). Thus, the prominent SOD isoform b was characterized as MnSOD and targeted for the further experiments.
Biochemical properties of H. armigera MnSOD
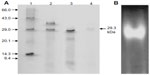
The identified MnSOD was partially purified to near-homogeneity using gel filtration chromatography. After gel filtration chromatography, the specific activity of purified enzyme was found to be 6,348.0 U/mg/min with 26.61% yield (Table 1). The purified enzyme appeared as single protein band with a molecular mass of ~29.3 kDa in SDS polyacrylamide gel (Figure 4A). Native PAGE of the same eluted fraction showed the single MnSOD isoform b (Figure 4B). Activity of MnSOD at different temperatures was checked by spectrophotometric assay. Enzyme activity was not changed up to 10°C. Further, the activity was changed with respect to change in temperature. Maximum activity was observed at 60°C. As temperature increased, the activity was further declined and came to its lowest peak at 90°C (Figure 5A). MnSOD activity was also investigated at different pH ranging from 2.0 to 12.0. H. armigera MnSOD showed maximum activity at pH 11.0 (Figure 5B).
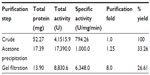  | Table 1 Purification of Helicoverpa armigera MnSOD |
Discussion
H. armigera has acquired strong resistance to various chemical pesticides and plant toxins, and thus, necessarily has developed a strong antioxidant mechanism.11,12 SODs are a foremost constituent of antioxidant system, and their activities are found to be increased in pesticide-resistant strains of some insect species.19,20
The present investigation reveals the presence of five SOD activity isoforms in the midgut of H. armigera. Insect polyphagy might be attributed to the existence of numerous and diverse antioxidant and detoxifying enzymes and their isoforms.32,33 Identification and upregulation of number of SOD activity isoforms in insects against various stresses have been reported previously.14,18 Studies confirmed the presence of different SOD activity isoforms in several insect species. Our results are consistent with previous findings reported in insect species. Interestingly, in the present study, total SOD activity was observed to be increased continuously as the larval developmental stages progressed. Maximum enhancement in the total SOD activity was observed in fourth instar, and the activity was further declined at fifth to sixth instar of the larvae. At these stages, larvae may attempt to do vigorous feeding, and thus might have chance to encounter several phytochemicals. Vigorous feeding may lead to increased metabolic rate and consequently formation of more ROS, which can result in an accelerated antioxidant mechanism.
SODs are ubiquitous metalloenzymes isolated and characterized from various species of organisms.34 In eukaryotes, there are two distinct isoforms of SODs distinguished by the type of metal present at their active sites: CuSOD, ZnSOD, and MnSOD. We have checked the effect of various divalent metal ions on H. armigera total SOD activity. H. armigera total SOD activity was significantly increased in the presence of metal ions namely Mn, Cu, Zn, and Fe, while decreased in the presence of Pb and Ca. These outcomes suggest the need of divalent metal ions for SOD; however, this is the major distinguishing character of SODs for their classification.30,35 Naturally, metal ions serve many functions in proteins, including the modification of protein structures, enhancement of the structural stability of the proteins in the conformation required for biological function, or taking part in the catalytic processes of enzymes. For SOD enzymes, the presence of metal ions is crucial for activity.29,30 Each SOD isozyme has been found to have a particular characteristic sensitivity toward a number of reagents. Cyanide was found to inhibit the CuSOD and ZnSOD but not the MnSOD. The product of the dismutation reaction, hydrogen peroxide, inactivates both the Cu/Zn and Fe but not the MnSOD.29 Various methods have been developed to distinguish SODs, which are based on selective inhibition or reconstitution of a particular form of enzyme.30,36 Here, the major isoform b with the highest activity was characterized as MnSOD, since it was found insensitive to H2O2. Formerly, MnSOD was identified and characterized from the important insect species such as Bombyx mori and Drosophila melanogaster.12,37
The apparent molecular mass of purified SOD protein was ~29.3 kDa. The observed molecular mass of the H. armigera MnSOD is consistent with those of the MnSODs characterized so far in the insect species.37 The H. armigera MnSOD was found stable in a wide temperature range (20°C–90°C) under the experimental conditions, and its stability was similar to those of other insect SODs.37 Previous studies have shown that temperature stress can cause MnSOD induction in several species.38 An earlier study indicated that the activity of Cu/ZnSOD in Bemisia tabaci was noticeably enhanced under cold and heat stress.39 In another study, the authors found that the MnSOD activity in B. tabaci was also increased with the high temperature conditions.38 H. armigera MnSOD showed maximum activity at pH 11. Results are consistent with previously reported pH optima for SOD activity in Spodoptera litura.13 Gut pH conditions are likely to have a major influence on the efficiency of nutrient extraction in insect herbivores.40 Although some plant cells are macerated during the mechanical process of ingestion, most cell lysis occurs in the gut, and midgut alkalinity greatly enhances extraction of foliage proteins. At high pH, the stability of superoxide anion radical is of the order of 100,000-fold greater than at the mildly acidic pH of grasshopper.41 MnSOD activity found at alkaline pH could be more useful in catalyzing the dismutation of superoxide in the high pH midgut of H. armigera. A detailed biochemical and molecular analysis of different SOD activity isoforms of H. armigera upon exposure to a particular stress will highlight their specific roles.
Acknowledgment
PRL acknowledges the support from the Department of Biotechnology, Government of India, New Delhi, for Research Associateship.
Disclosure
The authors report no conflicts of interest in this work.
References
Felton GW, Summers CB. Antioxidant systems in insects. Arch Insect Biochem Physiol. 1995;29:187–197. | |
Aucoin RR, Philogene BJR, Arnason JT. Antioxidant enzymes as biochemical defenses against phototoxin-induced oxidative stress in three species of herbivorous Lepidoptera. Arch Insect Biochem Physiol. 1991;16:139–152. | |
Wang Y, Oberley LW, Murhammer DW. Antioxidant defense systems of two Lepidopteran insect cell lines. Free Radic Biol Med. 2001;30:1254–1262. | |
Weirich GF, Collins AM, Williams VP. Antioxidant enzymes in the honey bee, Apis mellifera. Apidologie. 2002;33:3–14. | |
Dubovskiy IM, Martemyanov VV, Vorontsova YL, Rantala MJ, Gryzanova EV, Glupov VV. Effect of bacterial infection on antioxidant activity and lipid peroxidation in the midgut of Galleria mellonella L. larvae (Lepidoptera, Pyralidae). Comp Biochem Physiol C. 2008;148:1–5. | |
Gujar GT, Kumar A, Kalia V, Chandrashekhar K. Spatial and temporal variation in susceptibility of American bollworm, Helicoverpa armigera (Hubner) to Bacillus thuringiensis var. Karstakin in India. Curr Sci. 2000;78:995–1001. | |
Rajapakse CNK, Walter GH. Polyphagy and primary host plants: oviposition preference versus larval performance in the lepidopteran pest Helicoverpa armigera. Arthropod Plant Interact. 2007;1:17–26. | |
Adamski Z, Ziemnicki K, Fila K, Zikic RV, Stajn A. Effects of long-term exposure to fenitrothion on Spodoptera exigua and Tenebrio molitor larval development and antioxidant enzyme activity. Biol Lett. 2003;40:43–52. | |
Bagchi D, Bagchi M, Hassoun EA, Stohs SJ. In vitro and in vivo generation of reactive oxygen species, DNA damage and lactate dehydrogenase leakage by selected pesticides. Toxicology. 1995;104:129–140. | |
Vontas JG, Small GJ, Hemingway J. Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem J. 2001;357:65–72. | |
Kannan K, Jain SK. Oxygen radical generation and endosulfan toxicity in Jurkat T-cells. Mol Cell Biochem. 2003;247:1–7. | |
Pritsos CA, Ahmad S, Elliott AJ, Pardini RS. Antioxidant enzyme level response to prooxidant allelochemicals in larvae of the southern armyworm moth, Spodoptera eridania. Free Radic Res Commun. 1990;9:127–133. | |
Ahmad S, Pardini RS. Mechanisms for regulating oxygen toxicity in phytophagous insects. Free Radic Biol Med. 1990;8:401–413. | |
Krishnan N, Kodrik D. Antioxidant enzymes in Spodoptera littoralis (Boisduval): are they enhanced to protect gut tissues during oxidative stress? J Insect Physiol. 2006;52:11–20. | |
Datkhile KD, Mukhopadhyaya R, Dongre TK, Nath BB. Increased level of superoxide dismutase (SOD) activity in larvae of Chironomus ramosus (Diptera: Chironomidae) subjected to ionizing radiation. Comp Biochem Physiol C. 2009;149:500–506. | |
Yamamoto K, Zhang P, He N, et al. Molecular and biochemical characterization of manganese-containing superoxide dismutase from the silkworm, Bombyx mori. Comp Biochem Physiol B. 2005;142:403–409. | |
Choi YS, Lee KS, Yoon HJ, Kim I, Sohn HD, Jin BR. Bombus ignitus Cu, Zn superoxide dismutase (SOD1): cDNA cloning, gene structure, and up-regulation in response to paraquat, temperature stress, or lipopolysaccharide stimulation. Comp Biochem Physiol B. 2006;144:365–371. | |
Wang Y, Wang L, Zhu Z, Mab W, Lei C. The molecular characterization of antioxidant enzyme genes in Helicoverpa armigera adults and their involvement in response to ultraviolet-A stress. J Insect Physiol. 2012;58:1250–1258. | |
Nardini L, Christian RN, Coetzer N, Koekemoer LL. DDT and pyrethroid resistance in Anopheles arabiensis from South Africa. Parasit Vectors. 2013;6:229. | |
Mamidala P, Wijeratne AJ, Wijeratne S, et al. RNA-Seq and molecular docking reveal multi-level pesticide resistance in the bed bug. BMC Genomics. 2012;13:6. | |
Nagarkatti S, Prakash A. Rearing Helicoverpa armigera (Hubner) on an artificial diet, Volume 17; 1974. In: Technical Bulletin, Commonwealth Institute of Biological Control, Bangalore. | |
Lomate PR, Jadhav BR, Giri AP, Hivrale VK. Alterations in the Helicoverpa armigera midgut digestive physiology after ingestion of pigeon pea inducible leucine aminopeptidase. PLoS One. 2013;8(9):e74889. | |
Pauchet A, Muck A, Svatos A, Heckel DG, Prelss S. Mapping the larval midgut lumen proteome of Helicoverpa armigera, a generalist herbivorous insect. J Proteome Res. 2007;7:1629–1639. | |
Lomate PR, Hivrale VK. Effect of Bacillus thuringiensis (Bt) Cry1Ac toxin and protease inhibitor on growth and development of Helicoverpa armigera (Hübner). Pest Biochem Physiol. 2013;105:77–83. | |
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:453–464. | |
Davis BJ. Disc electrophoresis II: methods and application to human serum. Ann N Y Acad Sci. 1964;12:404–427. | |
Chen C, Pan S. Assay of superoxide dismutase activity by combining electrophoresis and densitometry. Bot Bull Acad Sin. 1996;37:107–111. | |
Imanari T, Hirota M, Miyazaki M, Hayakawa K, Tamura Z. Improved assay method for superoxide dismutase. Igaku no ayumi. 1997;101:496–497. | |
Hodgson EK, Fridovich I. The interaction of bovine erythrocyte superoxide dismutase with hydrogen peroxide: inactivation of the enzyme. Biochemistry. 1975;14:5294–5299. | |
Bannister JV, Bannister WH, Rotilio G. Aspects of the structure, function, and applications of superoxide dismutase. CRC Crit Rev Biochem. 1987;22:111–180. | |
Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. | |
Dawkar VV, Chikte YR, Lomate PR, Dholakia BB, Gupta VS, Giri AP. Molecular insights in to defense mechanisms of Lepidopteron insect pests against toxicants. J Proteome Res. 2013;12:4727–4737. | |
de la Paz Celorio-Mancera M, Wheat CW, Vogel H, Söderlind L, Janz N, Nylin S. Mechanisms of macroevolution: polyphagous plasticity in butterfly larvae revealed by RNA-Seq. Mol Ecol. 2013;22:4884–4895. | |
Fridovich I. Superoxide dismutase. Adv Enzymol. 1986;58:61–97. | |
Lee YM, Ayala FJ, Misra HP. Purification and properties of superoxide dismutase from Drosophila melanogaster. J Biol Chem. 1981;256:8506–8509. | |
Asada K, Kanematsu S, Okada S, Hayakawa T. Phylogenetic distribution of three types of superoxide dismutase in organisms and in cell organelles. In: Bannister JV, Hill HAO, editors. Chemical and Biochemical Aspects of Superoxide and Superoxide Dismutase. Vol IIA. New York: Elsevier; 1980:136. | |
Duttaroy A, Meidinger R, Kirby K, Carmichael S, Hilliker A, Phillips J. A manganese superoxide dismutase-encoding cDNA from Drosophila melanogaster. Gene. 1994;143:223–225. | |
Gao XL, Li JM, Wang YL, et al. Cloning, expression and characterization of mitochondrial manganese superoxide dismutase from the whitefly, Bemisia tabaci. Int J Mol Sci. 2013;14:871–887. | |
Li JM, Su YL, Gao XL, He J, Liu SS, Wang XW. Molecular characterization and oxidative stress response of an intracellular Cu/Zn superoxide dismutase (CuZnSOD) of the whitefly, Bemisia tabaci. Arch Insect Biochem. 2011;77:118–133. | |
Appel HM. The chewing herbivore gut lumen: physicochemical conditions and their impact on plant nutrients, allelochemicals and insect pathogens. In: Bernays EA, editor. Insect-Plant Interactions. Vol V. Boca Raton, FL: CRC Press; 1994:209–223. | |
Rosen GM, Britigan BE, Halpern HJ, Pou S. Free Radicals: Biology and Detection by Spin Trapping. Oxford: Oxford University Press; 1999. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.