Back to Journals » OncoTargets and Therapy » Volume 13
STRN-ALK Fusion in Lung Adenocarcinoma with Excellent Response Upon Alectinib Treatment: A Case Report and Literature Review
Authors Su C, Jiang Y, Jiang W, Wang H, Liu S, Shao Y, Zhao W, Ning R, Yu Q
Received 19 September 2020
Accepted for publication 6 November 2020
Published 4 December 2020 Volume 2020:13 Pages 12515—12519
DOI https://doi.org/10.2147/OTT.S282933
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Cuiyun Su,1 Ya Jiang,2 Wei Jiang,1 Huilin Wang,1 Sisi Liu,2 Yang Shao,2 Wenhua Zhao,1 Ruiling Ning,1 Qitao Yu1
1Department of Respiratory Oncology, Guangxi Medical University Affiliated Tumor Hospital, Nanning, China; 2Department of Research and Development, Nanjing Geneseeq Technology Inc, Nanjing, Jiangsu, China
Correspondence: Qitao Yu Email [email protected]
Abstract: Non-small cell lung cancer (NSCLC) patients with anaplastic lymphoma kinase (ALK) rearrangement benefit from treatment with ALK inhibitors. Therefore, the identification of druggable ALK fusions is necessary for NSCLC treatment. More than 90 fusion partners of ALK have been reported in NSCLC patients, but the striatin gene (STRN)-ALK fusion has rarely been reported. Moreover, the response of STRN-ALK fusion patients treated with ALK inhibitors remains to be explored. A 64-year-old Chinese male with no history of smoking or alcohol consumption was diagnosed as stage IVB lung adenocarcinoma (LADC) (cT4N3M1c) in October 2018. Next-generation sequencing (NGS) targeting 425 cancer-related genes was performed on the plasma and supernatant of pleural effusion samples and revealed an STRN-ALK fusion. The patient received alectinib (600 mg, twice daily) as the first-line treatment with an excellent response exceeding 19 months. This is the first report of a NSCLC patient harboring an STRN-ALK fusion and exhibiting an excellent response to alectinib treatment. This case provides valuable information for therapeutic decision-making of patients with STRN-ALK fusions. Furthermore, this case also highlighted the advantage of performing targeted NGS on circulating tumor DNA for the identification and analysis of rare, druggable genomic alterations.
Keywords: lung adenocarcinoma, STRN-ALK fusion, ctDNA, alectinib, targeted NGS
Introduction
Chromosome rearrangements involving anaplastic lymphoma kinase (ALK) have been identified as drug-targetable gene alterations in non-small cell lung cancer (NSCLC) patients.1 NSCLC patients with ALK rearrangements benefit from ALK-tyrosine-kinase-inhibitors (ALK-TKIs), such as crizotinib (first-generation), ceritinib, alectinib, brigatinib (second-generation), lorlatinib (third-generation), and others under development (eg, ensartinib, entrectinib, repotrectinib).2–4 Therefore, the identification of druggable ALK fusions is crucial for developing effective treatment strategies in NSCLC patients.5
Compared to traditional methods, next-generation sequencing (NGS) provides more comprehensive details of genomic alterations in cancer patients, and is advantageous in revealing novel ALK fusions.6 By the end of January 2020, more than 90 ALK fusion partners have been reported in NSCLC patients;7 however, the striatin (STRN)-ALK fusion has rarely been reported.8 Additionally, the efficacy of targeting rare ALK fusions using different ALK-TKIs is not sufficiently clear. The case of a 51-year-old male NSCLC patient harboring the STRN-ALK fusion, without ALK resistance mutations, failed to exhibit a response to alectinib treatment.9 In this study, we describe a patient with stage IVB lung adenocarcinoma who harbored a STRN-ALK fusion and exhibited an excellent response to alectinib treatment.
Case Presentation
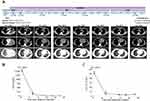
A 64-year-old Chinese male with no history of smoking or alcohol consumption visited our hospital in October 2018 with the chief complaint of dyspnea and pain in his right leg and left chest. Computed tomography (CT) revealed a massive shadow in the upper lobe of the left lung, as well as pleural effusion, and metastases in both lungs and mediastinal lymph nodes (Figure 1A). Multiple bone metastases were also observed on a bone emission computed tomography (ECT) scan. No brain metastasis was detected. The cytology of the pleural effusion suggested stage IVB lung adenocarcinoma (LADC) (cT4N3M1c). Tumor markers carcinoembryonic antigen (CEA) and cancer antigen 125 (CA-125) were detected at high levels of 1500 ng/mL and 126.7 U/mL, respectively. The Eastern Cancer Cooperative Group (ECOG) performance status (PS) score for the patient was 2–3.
To establish an efficient therapeutic strategy, cell-free DNA from 4 mL of plasma and the supernatant of the pleural effusion were subjected to targeted NGS. Capture-based targeted sequencing was performed using a panel consisting of all exons and critical introns in 425 cancer-related genes spanning 1.46 MB of the human genome. The circulating tumor DNA (ctDNA) from plasma and pleural effusion revealed an STRN: exon3-ALK: exon20 fusion (Figure 2), with a high mutant allele frequency (8.7% and 15.9%, respectively). Other cancer-related gene mutations were also observed in plasma and pleural effusion samples, including SETD2 and GRM8 (Table 1). The patient had a low tumor mutation burden (2.2 mutation/Mb) and was microsatellite-stable. Immunohistochemistry analyses revealed negative expression for programmed death ligand 1 (PD-L1).
 |
Table 1 Genetic Alterations Detected in the Plasma and the Supernatant of Pleural Effusion |
The patient then underwent lumbar cement vertebroplasty in November 2018. At the same time, alectinib was administered at a dose of 600 mg, twice daily. Two weeks post-treatment, chest-CT showed a decreased tumor mass (Figure 1A), and the clinical ECOG PS was 1. Three months later, the patient achieved a partial response (PR), which was demonstrated by CT reexamination (Figure 1A), and a sustained response was ongoing as of April 2020.
Throughout the course of alectinib treatment, the levels of circulating CEA and CA-125 dramatically decreased and was maintained at a relatively low level compared to pre-treatment levels (Figure 1B and C). In May 2020, repetitive ctDNA analysis of targeted NGS failed to detect the STRN-ALK fusion or other mutations (Table 1), reflecting the real-time tumor response to alectinib therapy. Currently, the patient is still receiving alectinib treatment with good tolerance.
Discussion
Multiple ALK fusion types have been reported in NSCLC patients, among which, different fusion partners may affect the response to ALK-TKIs.1,10 Therefore, determinations of the sensitivity of different ALK fusions to different ALK inhibitors is necessary for therapeutic decision-making purposes. The STRN-ALK fusion is an extremely rare ALK rearrangement in NSCLC patients that was first described in 2013 using RNA sequencing.11 A literature review identified only two patients that harbored STRN-ALK fusions, and who had been treated with different ALK-TKIs and exhibited different responses (Table 2). One of the patients responded to crizotinib treatment after first- and second-line chemotherapy failure, and achieved a response for 6 months.8 The other case showed no response to alectinib treatment, which may have been due to the expression of vimentin and ABCB1, as they were reported to be potential drug-resistance factors against ALK-TKIs.9,12,13 Interestingly, all three reported cases (including the present case) were male NSCLC patients, indicating that the frequency of STRN-ALK fusions may be higher in males. Additional studies are required to verify this hypothesis.
 |
Table 2 Details of All Published Cases of NSCLC Patients with STRN~ALK Fusions Treated with ALK-TKIs |
The ALEX trial compared alectinib to crizotinib as first-line treatments for ALK-positive advanced NSCLC patients and identified a significantly prolonged progression-free survival (PFS) in patients treated with alectinib versus those treated with crizotinib (median PFS, 25.7 months vs 10.4 months, respectively).3,14–16 In 2018, alectinib was approved by the Food and Drug Administration (FDA) for the first-line management of ALK-rearranged NSCLC patients. Those clinical studies also indicated that patients with STRN-ALK fusions may have superior efficacy upon alectinib treatment. In our case, we chose to administer alectinib to the patient and an excellent benefit was obtained.
Repetitive ctDNA analysis revealed that the clearance of the plasma STRN-ALK fusion after alectinib treatment was associated with an excellent response and may have potential in predicting the efficacy of ALK-TKIs. To clearly understand the potential for alectinib to treat tumors bearing STRN-ALK fusions, the prevalence of this rearrangement needs to be assessed in more patients. At present, the patient is still receiving alectinib treatment and the response time has exceeded 19 months.
This case also highlighted the advantage of NGS for the identification of rare druggable genomic alterations and distinct genomic features. In our patient, targeted NGS of ctDNA from plasma and pleural effusion biopsies revealed similar molecular features as the patient was positive for the STRN-ALK fusion, but negative for other driver alterations.17 SETD2 E1553K and GRM8 E508K mutations were also identified at a relatively high mutant allele frequency.
SETD2 was reported to be one of the most frequently mutated genes in ALK-fusion-positive NSCLC patients, following that of TP53 and ALK mutations.17 Furthermore, a previous study also showed that SETD2 inactivation was cooperative with fusion oncogenes in TP53-wild-type LADCs,18 which was consistent with the features of the case reported here. However, targeted therapies against SETD2 inactivation have not yet been developed.19
It was reported that the components of the GRM8 signaling pathway could serve as potential targets for squamous cell lung cancer patients carrying GRM8 activating variants.20 However, the roles of GRM8 and its mutations on the progression of LADC remains unclear. In the present case, alectinib treatment obtained an excellent benefit, with the response time exceeding 19 months. This patient’s durable response to alectinib and future resistance mechanisms need to be followed-up.
Conclusion
In summary, by using targeted NGS-based ctDNA profiling in a clinical setting, we were able to detect a rare STRN-ALK fusion in a patient with advanced NSCLC. To our knowledge, this is the first report of a NSCLC patient with STRN-ALK fusion to exhibit an excellent response to alectinib treatment. This study also provides meaningful insight into the potential treatment options of lung adenocarcinoma patients with STRN-ALK fusions. Further studies verifying the sensitivity of different ALK fusion variants to alectinib treatment are necessary to aid in therapeutic decision-making.
Data Sharing Statement
All datasets generated for this study are included in the manuscript.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Guangxi Medical University Affiliated Tumor Hospital. The patient provided written informed consent to participate in this study. Written informed consent for the publication of this case report and accompanying images was obtained from the patient. We would like to thank the patient and his family for providing consent for publication. We also thank all staff involved in this case study.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This research did not receive any specific funding.
Disclosure
YJ, SL and YS are employees of Nanjing Geneseeq Technology Inc., China. YJ, SL and YS report personal fees from Geneseeq Technology Inc., during the conduct of the study. The authors report no other potential conflicts of interest for this work.
References
1. Lin JJ, Zhu VW, Yoda S, et al. Impact of EML4-ALK variant on resistance mechanisms and clinical outcomes in ALK-positive lung cancer. J Clin Oncol. 2018;36(12):1199–1206. doi:10.1200/jco.2017.76.2294
2. Minari R, Gnetti L, Lagrasta CA, et al. Emergence of a HER2-amplified clone during disease progression in an ALK-rearranged NSCLC patient treated with ALK-inhibitors: a case report. Transl Lung Cancer Res. 2020;9(3):787–792. doi:10.21037/tlcr.2020.04.03
3. Young K, Toyoaki H, Hiroshi N, et al. MA07.03 Alectinib (ALC) versus Crizotinib (CRZ) in ALK-positive non-small cell lung cancer (ALK+ NSCLC): primary results from phase III study (J-ALEX). J Thoracic Oncol. 2017;12(1):S378–S379. doi:10.1016/j.jtho.2016.11.427
4. Facchinetti F, Tiseo M, Di Maio M, et al. Tackling ALK in non-small cell lung cancer: the role of novel inhibitors. Transl Lung Cancer Res. 2016;5(3):301–321. doi:10.21037/tlcr.2016.06.10
5. Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med. 2020;12(1):8. doi:10.1186/s13073-019-0703-1
6. Vendrell JA, Taviaux S, Béganton B, et al. Detection of known and novel ALK fusion transcripts in lung cancer patients using next-generation sequencing approaches. Sci Rep. 2017;7(1):12510. doi:10.1038/s41598-017-12679-8
7. Ou S-HI, Zhu VW, Nagasaka M. Catalog of 5ʹ Fusion Partners in ALK-positive NSCLC Circa 2020. JTO Clinical Research Reports. 2020;1(1):100015. doi:10.1016/j.jtocrr.2020.100015
8. Yang Y, Qin SK, Zhu J, et al. A rare STRN-ALK fusion in lung adenocarcinoma identified using next-generation sequencing-based circulating tumor DNA profiling exhibits excellent response to crizotinib. Mayo Clinic Proc Innovations, Quality Outcomes. 2017;1(1):111–116. doi:10.1016/j.mayocpiqo.2017.04.003
9. Nakanishi Y, Masuda S, Iida Y, Takahashi N, Hashimoto S. Case report of non-small cell lung cancer with STRN-ALK translocation: a nonresponder to alectinib. J Thorac Oncol. 2017;12(12):e202–e204. doi:10.1016/j.jtho.2017.08.009
10. Yoshida T, Oya Y, Tanaka K, et al. Differential crizotinib response duration among ALK fusion variants in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2016;34(28):3383–3389. doi:10.1200/jco.2015.65.8732
11. Majewski IJ, Mittempergher L, Davidson NM, et al. Identification of recurrent FGFR3 fusion genes in lung cancer through kinome-centred RNA sequencing. J Pathol. 2013;230(3):270–276. doi:10.1002/path.4209
12. Urbanska EM, Sørensen JB, Melchior LC, Costa JC, Santoni-Rugiu E. Changing ALK-TKI-resistance mechanisms in rebiopsies of ALK-rearranged NSCLC: ALK- and BRAF-mutations followed by epithelial-mesenchymal transition. Int J Mol Sci. 2020;21(8):2847. doi:10.3390/ijms21082847
13. Gainor JF, Dardaei L, Yoda S, et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov. 2016;6(10):1118–1133. doi:10.1158/2159-8290.cd-16-0596
14. Peters S, Camidge DR, Shaw AT, et al. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med. 2017;377(9):829–838. doi:10.1056/NEJMoa1704795
15. Hida T, Nokihara H, Kondo M, et al. Alectinib versus crizotinib in patients with ALK-positive non-small-cell lung cancer (J-ALEX): an open-label, randomised Phase 3 trial. Lancet. 2017;390(10089):29–39. doi:10.1016/s0140-6736(17)30565-2
16. Gridelli C, Casaluce F, Sgambato A, Palazzolo G, Maione P. J-ALEX trial will crown alectinib as the standard choice for anaplastic lymphoma kinase positive untreated non-small cell lung cancer patients? J Thorac Dis. 2018;10(1):106–108. doi:10.21037/jtd.2017.12.76
17. Liu S, Huang T, Liu M, et al. The genomic characteristics of ALK fusion positive tumors in chinese NSCLC patients. Front Oncol. 2020;10:726. doi:10.3389/fonc.2020.00726
18. Lee JJ, Park S, Park H, et al. Tracing oncogene rearrangements in the mutational history of lung adenocarcinoma. Cell. 2019;177(7):1842–1857.e21. doi:10.1016/j.cell.2019.05.013
19. Walter DM, Venancio OS, Buza EL, et al. Systematic in vivo inactivation of chromatin-regulating enzymes identifies setd2 as a potent tumor suppressor in lung adenocarcinoma. Cancer Res. 2017;77(7):1719–1729. doi:10.1158/0008-5472.can-16-2159
20. Zhang P, Kang B, Xie G, et al. Genomic sequencing and editing revealed the GRM8 signaling pathway as potential therapeutic targets of squamous cell lung cancer. Cancer Lett. 2019;442:53–67. doi:10.1016/j.canlet.2018.10.035
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.