Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 13
STEAP4 Inhibits HIF-1α/PKM2 Signaling and Reduces High Glucose-Induced Apoptosis of Retinal Vascular Endothelial Cells
Authors Liu L, Xu H, Zhao H, Jiang C
Received 27 February 2020
Accepted for publication 29 April 2020
Published 20 July 2020 Volume 2020:13 Pages 2573—2582
DOI https://doi.org/10.2147/DMSO.S251663
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Juei-Tang Cheng
Lei Liu,1 Hui Xu,1 Hongyu Zhao,2 Chunying Jiang1
1Department of Ophthalmology, The First Hospital of Jilin University, Changchun 130021, People’s Republic of China; 2Department of Radiation Oncology, China-Japan Union Hospital of Jilin University, Changchun 130033, People’s Republic of China
Correspondence: Chunying Jiang
Department of Ophthalmology, The First Hospital of Jilin University, No. 71 Xinmin Street, Changchun 130021, People’s Republic of China
Email [email protected]
Background: Diabetic retinopathy (DR) is a vascular lesion induced by high glucose. STEAP4 is an indispensable membrane protein, which is closely related to hyperglycemic-induced cell inflammation and injury, while STEPT4 has not been studied in hyperglycemic-induced retinal vascular endothelial cell injury.
Methods: The expression of STEAP4 was detected by RT-qPCR and Western blot. CCK-8 was used to detect cell survival. STEAP4 was overexpressed by cell transfection. The expressions of cytokines TNF-α, IL-1, IL-6, ICAM-1, MDA, SOD and ROS were detected by ELISA. Cell apoptosis was detected by flow cytometry. The expressions of proteins associated with cell damage VEGF, KLF2, eNOS and apoptosis-related proteins Bax, cleaved caspase3 and Bcl2 were detected by Western blot. Finally, the expressions of HIFα and PKM2 were detected by immunofluorescence and Western blot.
Results: The expression of STEAP4 in hyperglycemic-induced retinal vascular endothelial cells (HRCECs) decreased gradually. Overexpression of STEAP4 reduced inflammation and apoptosis of HRCECs and improved dysfunction of them. Meanwhile, overexpression of steap4 inhibited the expression of HIF-1α/PKM2 signal.
Conclusion: STEAP4 can be a potential therapeutic target for diabetic retinopathy by inhibiting HIF1/PKM2 signaling to reduce hyperglycemic-induced retinal cell apoptosis.
Keywords: diabetic retinopathy, STEAP4, HIF-1α/PKM2, HG-induced retinal vascular endothelial cells
Introduction
Diabetic retinopathy (DR) is one of the most common vascular complications of diabetes, which is a metabolic disorder with vascular lesions induced by long-term and sustained high glucose.1 DR is the principal factor of visual dysfunction and blindness in people aged 20–65 worldwide. All over the world, the number of people with diabetes is increasing rapidly, and the prevalence of diabetes is increasing.
The pathogenesis of DR is not yet fully understood, and the factors influencing its occurrence and development are also complicated. Systemic factors leading to DR include age, course of disease, blood glucose, blood lipid, etc.2 Some researches hold the belief that upon the persistent stimulation of high sugar, the retina will produce a series of pathophysiological processes, including oxidative stress, inflammation, glycosylation end products gathered and neurologic abnormalities, and lead to retinal microvascular lesion and blood retinal barrier damage, thus eventually triggering macular edema and retinal neovascularization.3
Six-Transmembrane Epithelial Antigen of the Prostate 4 (STEAP4), also known as six-transmembrane protein of prostate 2 (STAMP2) or TNFα- induced adipose-related proteins (TIARP), is an indispensable membrane protein and anti-inflammatory protein.4 STEAP4 is located in the long arm of chromosome 7, an area known to be susceptible to type 2 diabetes, insulin resistance and rheumatism.5–7 In 2001, in a study on childhood obesity, STEAP4 was screened for differential expression in adipose tissue of obese and normal children. Obesity, as a metabolic disease caused by multiple factors, plays an important role in the occurrence and development of many chronic diseases, such as diabetes.8 Studies have shown that overexpression of STEAP4 can inhibit inflammation in streptozotocin-induced diabetic models of mouse and can better regulate glucose metabolism.9 Mice interfered withSTEAP4 regulated insulin resistance, hyperglycemia, and inflammatory.10 Inflammatory cytokines, hormones and other cellular stress indicators regulate the expression of STEAP4, which can protect cells from damage and help maintain normal metabolic function. Overexpression of STEAP4 can inhibit atherosclerosis in diabetic mice,11 and STEAP4 can reduce damage to renal mesangial cells induced by high glucose or S100B.12 However, STEAP4 has not been investigated in hyperglycemic-induced retinal vascular endothelial cell injury.
Therefore, the purpose of this study was to examine the effect of STEAP4 on retinal vascular endothelial cells exposed to HG conditions and to explore the mechanism.
Methods and Materials
Cell Culture
Human Retinal Capillary Endothelial Cells (HRCECs) were purchased from Type Culture Collection of the Chinese Academy of Science. Cells were cultured in DMEM (Gibco; Thermo Fisher Scientific) and 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific) with 5% CO2 at 37°C.
Cell Treatments
Cells were seeded in 6-well plates with 2 × 105 cells per well and then incubated for 12 h. When the cells were in good condition and grew logarithmically, the cells were collected after 12 or 24 or 48 hours of high glucose induction. The Control group was given the same dose of normal saline and the mannitol group (MA) was given the same dose of mannitol.
Real‐Time Quantitative Reverse Transcription–Polymerase Chain Receptor (qRT–PCR)
Total RNA was extracted with TRI zol reagent (Invitrogen, Inc.) under the guidance of the manufacturer’s instructions, and cDNA was synthesized using a reverse transcription kit (Invitrogen, Inc.). qRT–PCR was performed using TaqMan probe method with the 7500 real‐time PCR system (Applied Biosystems, Foster City, CA). The expression level of target genes expression level was normalized to GAPDH. Primer sequences were as follows: STEAP4 forward: 5′-GCGCCTCTCCCTCAGTTATG-3′; STEAP4 reverse: 5′-GGTCTTCTGGGGGTTTCGAC-3′. GAPDH forward: 5ʹ-AGCCACATCGCTCAGACAC-3ʹ; GAPDH reverse: 5ʹ-GCCCAATACGACCAAATCC-3ʹ. GAPDH was included as an internal control and 2−ΔΔCq method was used for statistical analysis.13
Western Blot
Cells were washed with PBS and lysed by RIPA lysis buffer (Thermo Fisher Scientific) and incubated for 30 min on ice. Fifteen micrograms of protein per lane was subjected to SDS-PAGE, and then transferred to a PVDF membrane. The membrane was incubated with primary antibody overnight at 4°C. The membrane was washed with PBS for 3 times the next day. After incubation with HRP-conjugated secondary antibody (1:5000; cat. no. AA24142) for 1 hour at room temperature. The signals were detected using enhanced chemiluminescence reagent (GE Healthcare) and Amersham Imager 600 (GE Healthcare Life Science). Anti- STEAP4 (1:1000; cat. no. PA5-20,407), anti- eNOS (1:1000; cat. no. PA1-037), anti-KLF2 (1:1000; cat. no. PA5-40,591), anti- ICAM-1 (1:1000; cat. no. MA1-80,910), anti-VEGF (1:1000; cat. no. MA5-13,182), anti-Bax (1:1000; cat. no. MA5-14,003), anti-Bcl-2 (1:1000; cat. no. MA5-11,757), anti-Cleaved-caspase3 (1:1000; cat. no. PA5-38,438), anti-Caspase3 (1:1000; cat. no. PA5-86,276), anti- HIF-1α (1:1000; cat. no. A700-001), anti-PKM2 (1:1000; cat. no. PA5-23,034) and anti-GAPDH (1:1000; cat. no. 5174S) antibodies were obtained from Thermo fisher technology.
Cell Counting Kit-8 (CCK-8) Assay
Cells were seeded into 96-well plates and incubated at 37°C in a 5% CO2 humidified incubator. Cell viability was determined using the CCK-8 reagent (Dojindo Molecular Technologies, Inc.), according to the manufacturer’s protocol. After 12, 24 and 48 h, 10 µL CCK-8 solution (Dojindo Molecular Technologies, Inc.) was added to each well for 4 h, the optical density was measured at 450 nm using a microplate reader.
Cell Transfection
Cells were seeded in 6-well plates with 2 × 105 cells per well and then incubated for 24 h. Full length transcript of STEAP4 was polymerase chain reaction (PCR) amplified from cDNA using PrimeSTAR HS DNA polymerase (Takara, Inc.). The PCR amplified product was inserted into the Kpn I and BamH I sites of pcDNA (Invitrogen, Inc.) vector, termed pcDNA/STEAP4. STEAP4 promoter region containing the p50 binding sites was PCR amplified from genomic DNA using PrimeSTAR HS DNA polymerase (Takara, Inc.) and inserted into the Kpn I and Hind III sites of the luciferase reporter pGL3-Basic (Promega, Madison, WI) vector, termed pGL3-STEPA4-promoter. A scrambled overexpression of NC was used as negative control for overexpression of STEAP4. And then the overexpression of STEAP4 and control were transfected into cells at a concentration of 20 nM with Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocols.
Reactive Oxygen Species (ROS) Assay
ROS levels of cells were detected using a fluorescent probe, 2′,7′-dichlorodihydrofluorescein (DCHF) (Sigma), which could be rapidly oxidized into the highly fluorescent 2′,7′-dichlorofluorescein (DCF) in the presence of intracellular reactive oxygen species (ROS). Fluorescence was monitored with a laser scanning confocal microscope (Leica, Germany) at 488 nm. The amount of ROS was quantified as the relative fluorescence intensity of DCF per cell in the scan area.
Enzyme Linked Immunosorbent Assay (ELISA) for Factors
The expressions of interleukin (IL)-6, tumor necrosis factor (TNF)-α, IL-1β, ICAM-1, MDA and SOD were evaluated using ELISA. The cell supernatant was centrifuged for 5–10 minutes at 4°C (5000g). The levels of TNF-a, IL-1β, IL-6, ICAM-1, MDA and SOD were detected by ELISA kit (BioSource International, Camarillo, CA, USA). All operations were carried out in strict accordance with ELISA kit instructions.
Flow Cytometry Analysis
Cells were collected and washed in PBS for 3 times, and then the cells were re-suspended in a 200 μL binding buffer, followed by staining with 5 μL PI (BestBio) for 10 min in the dark. Next, cells were stained with Annexin V-FITC (BestBio) for 10 min in the dark at room temperature. At last, the cell cycle distribution was analyzed via flow cytometry with FlowJo software (BD Bioscience).
Immunofluorescence
Cells on a sheet of glass were fixed with 4% paraformaldehyde (PFA) for 30 min and then washed with PBS twice. Next, cells were permeated with PBS containing 0.1% Triton X-100 for 5 min. The cells were incubated with 5% skim milk powder to block non-specific staining. After 12 h of incubation with primary antibody at 4°C, the cells were washed three times with PBS. Later on, the samples were incubated with 10% goat serum at room temperature for 1 h. And then cells were further stained with fluorophore (Alex488 and Alexa 530)-conjugated secondary antibodies. After staining, the cells were counterstained with DAPI. The sealed slides were analysed using a Leica TCS-SP microscope with companion software.
Statistical Analysis
Statistical data analysis was performed with SPSS 22.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, Inc.). All experimental results are expressed as mean ± standard deviation. And data were analyzed using either ANOVA followed by a Tukey’s post hoc test for comparison of multiple groups or an independent Student’s t-test for comparison of two groups. P<0.05 was considered to indicate statistically significant differences. Data were obtained from at least three individual experiments.
Results
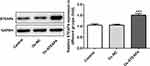
The Expression of STEAP4 in HG-Induced HRCECs Gradually Decreased
The sugar-induced concentration of the normal group was 5.5mmol/L, the mannitol induced concentration was 30mol/L and the HG-induced concentration was 30mol/L. After induction for 12h, 24h and 48h, the expression of STEAP4 was detected by RT-qPCR and Western blot. The results are shown in Figure 1A and B, compared with the normal and MA group, the expression of STEAP4 was significantly decreased in the HG-induced group. Moreover, the expression of STEAP4 in HRCECs gradually decreased with the time of high glucose stimulation, which was time-dependent. Then, CCK-8 was used to further detect the effect of HG on cell survival. We found that compared with HG induction for 24h, there was no significant change in survival after HG induction for 48h (Figure 1C). Therefore, high glucose induction at the concentration of 30mol/L and induction time of 48h were selected as the following experimental induction conditions.
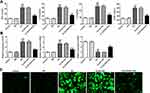
Overexpression of STEAP4 Attenuated the Inflammatory and Oxidative Stress Induced by HG
The overexpressed plasmid of STEAP4 was constructed and transfected into HRCECs induced by HG and the expression of STEAP4 detected by Western blot significantly increased (Figure 2). Compared with the control and MA group, the expression levels of TNF-α, IL-1β, IL-6, ICAM-1 in the HG–induced group were obviously increased. Compared with Overexpression-NC+HG group, TNF-α, IL-1β, IL-6, ICAM-1 decreased in Overexpression-STEAP4+HG group (Figure 3A). Subsequently, we detected the expression levels of ROS, MDA and SOD and found that compared with the control and MA group, the ROS and MDA expressions in the HG-induced group significantly increased. Compared with Overexpression-NC+HG, the ROS and MDA expressions of Overexpression-STEAP4+HG decreased, while the SOD expression showed the opposite trend (Figure 3B and C). These results suggest that overexpression of STEAP4 attenuates HG-induced inflammatory and oxidative stress responses in HRCECs.
Overexpression of STEAP4 Ameliorated HG-Induced Endothelial Dysfunction
To further test the effect of STAPT4 on HG-induced cell damage, we detected the expression of the proteins related to cell damage, including VEGF, KLF2, eNOS and ICAM-1 after the overexpression of STEAP4. As shown in Figure 4, compared with the control and MA group, the expression of KLF2 and eNOS in the HG-induced group decreased, while the expression of VEGF and ICAM-1 increased. Compared with Overexpression-NC +HG group, Overexpression-STEAP4+HG group showed significantly increased expression of KLF2 and eNOS, while decreased expression of VEGF and ICAM-1. Meanwhile, the expression of VEGF was detected by immunofluorescence, and the results were consistent with those obtained by Western blot (Figure 5). These results indicated that overexpression of STAEAP4 ameliorated HG-induced dysfunction of HRCECs.
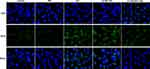 |
Figure 5 Overexpression of STEAP4 ameliorated HG-induced endothelial dysfunction. The expression of VEGF was detected by immunofluorescence. n=3. |
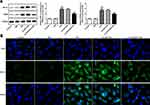
Overexpression of STEAP4 Decreased Apoptosis of HRCECs Induced by HG
Subsequently, apoptosis was measured and we found that compared with the control and MA group, the rate of apoptosis increased significantly in the HG-induced group (Figure 6A and B), accompanied by increased expression of apoptotic proteins Bax, cleaved caspase3 and decreased expression of Bcl2 (Figure 7). Compared with Overexpression-NC+HG group, the Overexpression-STEAP4+HG group showed significantly decreased apoptosis, decreased expression of Bax and cleaved caspase3, and increased expression of Bcl2. The results showed that STEAP4 could reduce apoptosis of HRCECs induced by HG.
Overexpression of STEAP4 Inhibited the Activity of HIF-1α/PKM2 Signal Pathway
To investigate the mechanism by which STEAP4 reverses HG-induced cell damage and apoptosis, we detected the expression of related proteins in the HIF-1α/PKM2 signaling pathway. As shown in Figure 8, we found that the HIF-1α/PKM2 expression level in the HG-induced group was significantly increased compared with that in the control and MA group. Compared with Overexpression-NC+HG group, the expression level of HIF-1α/PKM2 in Overexpression-STEAP4+HG group was obviously decreased. From this, we preliminarily concluded that STAPT4 reverses cell injury and apoptosis induced by HG through the HIF-1α/PKM2 signaling pathway.
Discussion
The damage of retinal vascular endothelial cell is an early marker of DR, and increased blood glucose is the main cause of DR. Diabetic patients show high blood glucose, which can induce retinal endothelial cell apoptosis. Studies have shown that retinal barrier destruction is caused by increased permeability of human retinal endothelial cells induced by high glucose.14 Duan et al also demonstrated that high glucose can induce inflammation, apoptosis and endothelial cell injury of vascular endothelial cells.15 In this study, high glucose concentration of 30mol/L was used to induce HRCECs, and we found that cell inflammation and oxidative stress response occurred, while high glucose could induce apoptosis and functional impairment of HRCECs. In our experiment, it was found that after HG induction, the cells developed inflammatory and oxidative stress responses, and endothelial cells suffered from functional damage.
STEAP4, as a six-fold transmembrane protein, has been demonstrated to be located on cell membranes, endosomal membranes, and Golgi membrane. In 2007, Moreno-Navarrete et al found that TNF-induced adipose-related protein (TIARP) induced by STEAP4 is a negative regulator of inflammation and insulin resistance. With normal diet, STEAP4 knockout mice could spontaneously form metabolic diseases, mainly manifested as insulin resistance, abnormal glucose tolerance and fatty liver. Whereas, insulin resistance plays a key role in the development of obesity-related diseases such as type 2 diabetes. At the same time, STEAP4 gene is located in 7q21.12, which is known to be susceptible to type 2 diabetes and insulin resistance. Decreased expression of STEAP4 can lead to dysfunction of visceral adipose tissue in obese diseases, mainly type 2 diabetes.16 Mice with low expression of STEAP4-regulated insulin resistance, hyperglycemia, and inflammation.10 In addition, overexpression of STEAP4 inhibited atherosclerosis in diabetic mice.11 STEAP4 can reduce the damage of renal mesangial cells induced by high glucose or S100B.12 Therefore, it is reasonable to speculate that STEAP4 gene plays an important role in retinal endothelial cell injury induced by high glucose. And our results confirmed our speculation. We found that the expression of STEAP4 gene was significantly increased in HG-induced HRCECs. Subsequently, after overexpression of STEAP4, we found that the expression level of cell inflammation and oxidative stress was increased, the level of was increased, and cell function was injured and apoptosis occurred.
Hypoxia‐inducible factor‐1α (HIF‐1α) is an important regulator of oxygen balance that plays a significant role in glycolysis and angiogenesis.17 As one of the important rate-limiting enzyme in energy metabolism, Pyruvate kinase M2 (PKM2) also plays an important role in the glycolytic pathway. PKM2 promotes the activation of related gene of glucose metabolism in cancer cells.18,19 In our experiments, the expressions of HIF‐1α and PKM2 in cells significantly increased in hyperglycemic-induced HRCECs. The results showed that glycolysis pathway was affected after hyperglycemia induction, which resulted in the stimulation of HIF‐1α and PKM2 expression. PKM2 is both the target gene of HIF-1α and the regulatory protein of HIF-1α. It has been reported that insulin can increase the expression of HIF-1α and PKM2 in HepG2 and HL7702 cells, and down-regulation of HIF-1α can inhibit the expression of PKM2 and insulin-induced glucose consumption.20 PKM2-HIF-1α target genes are involved in glucose metabolism, resulting in an increase of glucose uptake and lactate secretion in cancer cells.21 Therefore, we concluded that HIF-1α/PKM2 signaling pathways play an important role in the development of diabetes; however, the specific role of HIF-1α/PKM2 in diabetic retinopathy caused by diabetes has not been studied. In addition, after reading through the literature, we found that HIF-1α/PKM2 can be a potential therapeutic target of disease. Xue -fu-Zhu-Yu decoction downgrades the expression of HIF-1α and VEGF by inhibiting PKM2 and so prevent retinal ischemia in rats, which suggested that HIF-1α/PKM2 signaling pathway plays a role in the retina-related diseases.22 And in obese individuals, the expression of HIF‐1α increased due to hypoxia and pro-inflammatory stimulation, leading to increased expression of visceral STEAP4 increase.23 Then, there is a question: whether STEAP4/HIF‐1α/PKM2 plays a role in retinal endothelial cell injury induced by high glucose? In this article, we found that after induction of HRCECs by HG, the expression of HIF‐1α and PKM2 rise sharply, with cell apoptosis and damage increase. However, after STEAP4 was overexpressed, the expressions of HIF‐1α and PKM2 decreased significantly, and the apoptosis and damage of the cells decreased, from which we concluded that STEAP4 reduced HG-induced injury and apoptosis of HRCECs by inhibiting HIF‐1α/PKM2 signaling pathway (Please see Figure 9 for the detailed information.).
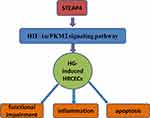 |
Figure 9 STEAP4 reduced HG-induced injury and apoptosis of HRCECs by inhibiting HIF‐1α/PKM2 signaling pathway. |
In conclusion, STEAP4 reduced HG-induced injury and apoptosis of HRCECs by inhibiting HIF‐1α/PKM2 signaling pathway and STEAP4/HIF‐1α/PKM2 can be a therapeutic target for DR.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Disclosure
The authors declare that they have no competing interests.
References
1. Cheung N, Mitchell P, Wong TY. Diabetic retinopathy. Lancet. 2010;376(9735):124–136. doi:10.1016/S0140-6736(09)62124-3
2. Jenkins AJ, Joglekar MV, Hardikar AA, et al. Biomarkers in diabetic retinopathy. Rev Diabet Stud. 2015;12(1–2):159–195. doi:10.1900/RDS.2015.12.159
3. Heng LZ, Comyn O, Peto T, et al. Diabetic retinopathy: pathogenesis, clinical grading, management and future developments. Diabet Med. 2013;30(6):640–650. doi:10.1111/dme.12089
4. Scarl RT, Lawrence CM, Gordon HM, et al. STEAP4: its emerging role in metabolism and homeostasis of cellular iron and copper. J Endocrinol. 2017;234(3):R123–R134. doi:10.1530/JOE-16-0594
5. Qi Y, Yu Y, Wu Y, et al. Genetic variants in six-transmembrane epithelial antigen of prostate 4 increase risk of developing metabolic syndrome in a Han Chinese population. Genet Test Mol Biomarkers. 2015;19(12):666–672. doi:10.1089/gtmb.2015.0104
6. Sharma PR, Mackey AJ, Dejene EA, et al. An islet-targeted genome-wide association scan identifies novel genes implicated in cytokine-mediated islet stress in type 2 diabetes. Endocrinology. 2015;156(9):3147–3156. doi:10.1210/en.2015-1203
7. Ebe H, Matsumoto I, Kawaguchi H, et al. Clinical and functional significance of STEAP4-splice variant in CD14 + monocytes in patients with rheumatoid arthritis. Clin Exp Immunol. 2018;191(3):338–348. doi:10.1111/cei.13076
8. Saxton SN, Clark BJ, Withers SB, et al. Mechanistic links between obesity, diabetes, and blood pressure: role of perivascular adipose tissue. Physiol Rev. 2019;99(4):1701–1763. doi:10.1152/physrev.00034.2018
9. Berner A, Bachmann M, Bender C, Pfeilschifter J, Christen U, Mühl H. Though active on RINm5F insulinoma cells and cultured pancreatic islets, recombinant IL-22 fails to modulate cytotoxicity and disease in a protocol of streptozotocin-induced experimental diabetes. Front Pharmacol. 2015;6:317.
10. Chen X, Huang Z, Zhou B. STEAP4 and insulin resistance. Endocrine. 2014;47(2):372–379.
11. Wang J, Han L, Wang Z-H, et al. Overexpression of STAMP2 suppresses atherosclerosis and stabilizes plaques in diabetic mice. J Cell Mol Med. 2014;18(4):735–748. doi:10.1111/jcmm.12222
12. Chuang C-T, Guh J-Y, Lu C-Y, et al. Steap4 attenuates high glucose and S100B-induced effects in mesangial cells. J Cell Mol Med. 2015;19(6):1234–1244. doi:10.1111/jcmm.12472
13. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
14. Jiao W, Ji J-F, Xu W, et al. Distinct downstream signaling and the roles of VEGF and PlGF in high glucose-mediated injuries of human retinal endothelial cells in culture. Sci Rep. 2019;9(1):15339. doi:10.1038/s41598-019-51603-0
15. Duan M-X, Zhou H, Wu -Q-Q, et al. Andrographolide protects against HG-induced inflammation, apoptosis, migration, and impairment of angiogenesis via PI3K/AKT-eNOS signalling in HUVECs. Mediators Inflamm. 2019;2019:6168340. doi:10.1155/2019/6168340
16. Moreno-Navarrete JM, Ortega F, Serrano M, et al. Decreased STAMP2 expression in association with visceral adipose tissue dysfunction. J Clin Endocrinol Metab. 2011;96(11):E1816–E1825. doi:10.1210/jc.2011-0310
17. Goda N, Kanai M. Hypoxia-inducible factors and their roles in energy metabolism. Int J Hematol. 2012;95(5):457–463. doi:10.1007/s12185-012-1069-y
18. Yang W, Xia Y, Hawke D, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2014;158(5):1210. doi:10.1016/j.cell.2014.08.003
19. Luo W, Hu H, Chang R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145(5):732–744. doi:10.1016/j.cell.2011.03.054
20. Li W, Wang J, Chen Q-D, et al. Insulin promotes glucose consumption via regulation of miR-99a/mTOR/PKM2 pathway. PLoS One. 2013;8(6):e64924. doi:10.1371/journal.pone.0064924
21. Wang H-J, Hsieh Y-J, Cheng W-C, et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1-mediated glucose metabolism. Proc Natl Acad Sci U S A. 2014;111(1):279–284. doi:10.1073/pnas.1311249111
22. Tan S-Q, Geng X, Liu J-H, et al. Xue-fu-Zhu-Yu decoction protects rats against retinal ischemia by downregulation of HIF-1alpha and VEGF via inhibition of RBP2 and PKM2. BMC Complement Altern Med. 2017;17(1):365. doi:10.1186/s12906-017-1857-2
23. Ozmen F, Ozmen MM, Gelecek S, et al. STEAP4 and HIF-1α gene expressions in visceral and subcutaneous adipose tissue of the morbidly obese patients. Mol Immunol. 2016;73:53–59. doi:10.1016/j.molimm.2016.03.008
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.