Back to Journals » OncoTargets and Therapy » Volume 12
Star Circular RNAs In Human Cancer: Progress And Perspectives
Authors Cheng Y, Sun H, Wang H, Jiang W, Tang W, Lu C, Zhang W, Chen Z, Lv C
Received 11 May 2019
Accepted for publication 20 September 2019
Published 7 October 2019 Volume 2019:12 Pages 8249—8261
DOI https://doi.org/10.2147/OTT.S215390
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yao Dai
Ye Cheng,1,* Hanzhi Sun,1,* Hanjin Wang,1,* Wei Jiang,1 Weiwei Tang,1 Chen Lu,1 Wenling Zhang,2 Ziyi Chen,3 Chengyu Lv1
1Department of General Surgery, Nanjing First Hospital, Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China; 2Department of Gastroenterology, Nanjing First Hospital, Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China; 3Department of General Surgery, Zhongda Hospital, Medical School, Southeast University, Nanjing, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chengyu Lv
Department of General Surgery, Nanjing First Hospital, Nanjing Medical University, Nanjing, Jiangsu, People’s Republic of China
Email [email protected]
Ziyi Chen
Department of General Surgery, Zhongda Hospital, Medical School, Southeast University, Nanjing, Jiangsu, People’s Republic of China
Email [email protected]
Abstract: Circular RNAs (circRNAs) are a recently discovered subclass of non-coding RNAs (ncRNAs) characterized by a covalently closed loop structure created by reverse splicing. Because they do not have a 5ʹ cap structure and a 3ʹ poly A tail, circRNAs have higher stability, abundance and evolutionary conservation than linear RNA between species. These features produce various potential biological functions of circRNAs, such as miRNA sponges, RNA-binding proteins that form RNA protein complexes. In recent years, more and more studies have shown that circRNAs play a vital role in the occurrence and development of human diseases. At the same time, their enormous potential as a biomarker and therapeutic target is also evolving. The purpose of this review is to summarize existing cancer-associated circRNAs and to try to find circRNAs that are abnormally expressed in many cancers. Therefore, we reviewed previous circRNAs studies related to cancer and selected them by statistics. The eight circRNAs that have the highest frequency in different cancers or involve key pathways are called star circRNAs. Here, we review the classification, features, and functions of emerging star circRNAs, with particular attention to the role of circRNAs in various cancers.
Keywords: circular RNA, cancer, targeted therapy, diagnosis
Introduction
In 1976, Sanger et al first proposed the concept of circular RNAs (circRNAs), and they found that some higher plant viruses are single-strand, covalently closed circRNA molecules.1 CircRNAs were first observed in humans in 1986 after infection with hepatitis D virus.2 Soon after, a group of researchers used the sensitivity analysis of RNA expression to find four circRNAs from the DCC gene in normal and tumor cells in rodents and humans.
CircRNAs are an endogenous RNA that can be formed between a downstream 3ʹ splice site and an upstream 5ʹ splice site in linear precursor mRNA (pre-mRNA).3,4 Due to technical deficiencies, these covalently closed circular RNA molecules are considered to be viroid-like, hepatitis virus molecule and splicing error results.5,6 With the advancement of bioinformatics tools, the identification of many circular RNAs has been revealed, and their important roles have been studied, such as Xu et al reviewed.7 In this review, we briefly describe the current understanding of the role of emerging star circRNAs and highlight their potential impact on cancer-targeted therapies.
Classification Of CircRNAs
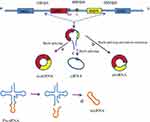
CircRNAs are produced by splice-mediated pre-sequence discontinuous splicing.8 This splicing process is completely different from traditional linear RNA splicing. According to the selection of splice sites, a single locus can splicing multiple circRNAs.9 According to the source sequence, circRNAs can be divided into four categories (Figure 1): a) exon circular RNAs (ecircRNAs), circRNAs originate from the exons of a linear transcript on the strand;10,11 b) exon-intron circular RNAs (EIciRNAs), circRNAs, circRNAs transcribe from the same gene positions of linear transcripts of adjacent genes;11 circular intron RNAs (ciRNAs), circRNAs originate from the intron of a linear transcript; c) circular intron RNAs (ciRNAs), circRNAs originate from the intron of a linear transcript;10,12 exon-intron circular RNAs (EIciRNAs), circRNAs, circRNAs transcribe from the same gene positions of linear transcripts of adjacent genes; and d) tRNA intronic circRNA(tricRNAs). tricRNAs derive from introns that are removed during pre-tRNA splicing,13,14 as Wu et al and Zhang et al reviewed.15,16 For these circRNAs, ecircRNAs account for approximately 85% of all identified circRNAs and are the most abundant type, and most of them are predominantly localized in the cytoplasm.10 In contrast, ciRNAs and EIciRNAs mainly locate in the nucleus indicate that these circRNAs have roughly different biological functions.
Biological Functions Of CircRNAs
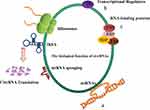
CircRNAs are considered to be a splicing by-product of long-term biological function, so past researchers have paid little attention to this nonlinear RNA. In recent years, more and more research has focused on circRNAs. Based on this, people have come to realize that circRNA might have many biological functions (Figure 2).17
CircRNAs Can Act As miRNA Sponges
Most circRNAs are primarily localized in the cytoplasm,10,18 and the results prompted researchers to study circRNA function in post-transcriptional regulation. MiRNAs can bind to their respective targets by bases paired with the 3ʹ-untranslated region (3ʹ-UTR) and act as a negative regulator of gene expression at the post-transcriptional level, thereby inhibiting protein translation.19
CircRNAs can bind miRNAs through a miRNA response element (MRE), which can act as a causative gene or a disease-promoting gene.10 The first identified human circRNA acts as a miRNA sponge, and the most powerful evidence for sponge activity is the antisense cerebellar degeneration-associated protein 1 transcript (CDR1as). CDR1as was first identified as a sponge of miR-7 in zebrafish neuron tissue, leading to midbrain developmental damage.20 Recent studies have shown that miR-7 plays an important role in the development and progression of cancer, respectively, and CDR1as as miR-7 sponge is important for all types of cancer. For example, the CDR1as/miR-7 signal axis may be a molecular target for the treatment of osteosarcoma.21 CDR1as may be a promising biomarker for hepatic microvascular invasion and a novel therapeutic target for inhibition of hepatocellular carcinoma (HCC) microvascular invasion.22
CircRNAs Can Function As Transcription Modulators
Recent studies have shown that circRNAs can act as a transcriptional regulator.23,24 For example, the intron circRNA ci-Ankyrin repeat domain 52 (ci-ankrd52) has been found to be enriched at its transcriptional site. Ci-ankrd52 may regulate its parental gene expression by modulating the prolonged activity of RNA polymerase II.21 The mechanism of action indicates that the circular intron transcripts exerts a new cis-regulatory effect on the expression of its parental coding gene. Further studies have shown that the EIciRNA-U1 small nuclear ribonucleoprotein (snRNP) complex is a special kind of circRNA, which may accelerate the expression of parental genes through RNA-RNA interaction, and complex with Pol II transcription on the parental gene promoter,24 as Ng, W. L et al reviewed.25
CircRNAs Can Interact With RBPs
RNA-binding proteins (RBPs) are a rich class of proteins involved in gene transcription and translation, and the basic elemental functions of circRNA, including occurrence, translation, target gene transcriptional regulation, and extracellular transport function through interaction with RBP.26 In addition, RBP can interact with circRNA and participate in splicing, processing, folding, stabilization, and localization of circRNA.27 For example, Dudekula et al uses a network tool called CircInteractome to identify has_circ_0000020 with multiple RBPs in the flanking sequence.28 The binding sites are on either side of the body sequence at a much higher frequency than the body sequences used to target the circRNA. This finding suggests that RBP tends to bind to the has_circ_0000020 intersection. Recent studies have shown that circRNAs can focus on specific components via RBP sponges, RBP assembly platforms and supertransporters, thereby competitively binding RBP, regulating RBP function and interacting with RBP.29,30
The Associations Between Star circRNAs And A Wide Range Of Diseases
By reference to previous circRNA-related literature, we have found that some circRNAs are involved in the development and progression of many diseases, and we call these circRNAs often appear in the literature as star circRNAs. We believe that by studying and analyzing star circRNA in cancer, we can explore the complete mechanism of circRNA-mediated cancer and use this mechanism to study whether circRNAs can develop into a clinically directed new tumor marker or tumor therapeutic target. Therefore, this article lists the eight most widely involved circRNA molecules in recent studies and briefly describes their function and related mechanisms in disease progression and progression, as well as the corresponding circRNAs as biomarkers or therapeutic targets. There are two broad categories between circRNAs and cancers: identifying potential biomarkers for cancer diagnosis by detecting circRNAs expression patterns and demonstrating the regulatory role of circRNAs in cancer development (Table 1).
 |  |  |  |
Table 1 A Summary Of Cancer Related circRNAs |
It is worth mentioning that there are many reasons for circRNA disorders in cancer, such as abnormal cis-elements, abnormal chromosomes and genomes, abnormal transcription, abnormal splice machinery and abnormal trans-acting factors. In addition, there are two hypothetical mechanisms for epigenetic aberrations involving circulator dysregulation: chromatin remodeling factors and post-translational modifications of histones affect transcription rate,31 and chromatin remodeling affects alternative splicing involving circRNAs biogenesis,4 as Wu et al previously commented on.15
Cerebellar Degeneration-Related Protein 1 Transcript Antisense RNA (CDR1as)
Hsa_circ_0001946, also known as CDR1as, is located at chrX: 139865339–139866824 with a total length of 1485. This gene represents naturally occurring RNA transcribed antisense with CDR1 (cerebellar degeneration associated protein 1, 34 kDa; GeneID: 1038), some of which may be circular rather than linear. Antisense circRNAs bind to miRNAs such as miR-7, acting as a sponge for these molecules and preventing them from interacting with target transcripts. The circRNA produced by this locus is down-regulated by miR-671, which also reduces the level of CDR1 transcript.
CDR1as is a circRNA molecule associated with the occurrence of various cancers and diseases. For instance, Xu B et al found that the CDR1as acting as miR-7 sponge could be the molecular target for the treatment of osteosarcoma;21 Jiang XM et al suggested that cCDR1as may serve as a potential vicious molecular biomarker to predict the aggressive tumor progression and worse prognosis for Cholangiocarcinoma (CCA) patients;32 Xu LL et al reported that CDR1as may be a promising biomarker of hepatic microvascular invasion and a novel therapy target for restraining microvascular invasion in HCC.33 In addition, the interaction between CDR1as and miR-7 has also been investigated in many non-tumor diseases. Xu HY et al revealed the effects of the strongly interacting pair of Cdr1as/miR-7 on insulin secretion;34 Li XB et al demonstrated that CDR1as acts as a miR-7 inhibitor, triggering the upregulation of GDF5 and subsequent Smad1/5/8 and p38 MAPK phosphorylation to promote osteogenic differentiation of periodontal ligament stem cells;35 and Yao WX et al indicated that the interaction between miR-7 and circRNA CDR1as may exert important functions and provide potential therapeutic targets in lung fibrotic diseases.36 Recent studies have shown that CDR1as can exert anti-oncogenic functions in bladder cancer by sponging miR-135a.37
Homeodomain Interacting Protein Kinase 3 (HIPK3)
The parental gene of circHIPK3 is located at 11p13 and contains a total of 19 exons. The gene HIPK3 is ubiquitous in fat (RPKM 29.7), gallbladder (RPKM 21.4) and 25 other tissues. HIPK3 is a protein-coding gene. Gene ontology (GO) annotations associated with this gene include transferase activity, transfer of phosphorus-containing groups, and protein tyrosine kinase activity.
Circ-HIPK3 is transcribed by the gene HIPK3 and is involved in tumorigenesis, invasion or metastasis of many cancers. At the same time, circ-HIPK3 may be a biomarker or therapeutic target for many cancers. Shuai MX et al indicated that circRNA_0000285(circ-HIPK3) may be a novel biomarker for nasopharyngeal carcinoma(NPC) and is involved in NPC radiosensitivity;38 Jin PC et al demonstrated that circ-HIPK3 contributes to glioma progression through targeting miR-654 from IGF2BP3 and implied circ-HIPK3 might be a potential target for glioma therapy;39 Kai D et al concluded that circHIPK3 promotes gallbladder cancer cell growth possibly by sponging miR-124 and the over-expressed circ-HIPK3 could be a novel therapeutic target and diagnosis marker of human gallbladder cancer;40 Ma XL et al suggested that circ-HIPK3 may become a novel potential biomarker for diagnosis and treatment target of osteocsarcoma.41
Itchy E3 Ubiquitin Protein Ligase (ITCH)
The coding gene for circ-ITCH is located at 20q11.22 and contains 31 exons. Gene ICTH is ubiquitously expressed in testis (RPKM 12.7), esophagus (RPKM 11.6) and 25 other tissues. The gene encodes a member of the Nedd4 family of the HECT structural domain E3 ubiquitin ligase. HECT domain E3 times in protein ligase transfers the ubiquitin from E2 times in protein conjugate to protein substrate, so as to target specific proteins for lysosomal degradation. Mutations in this gene are the cause of multiple autoimmune syndromes. A variety of spliced transcripts of the same type encoding this gene have been observed.
Circ-ITCH acts primarily as a miRNA sponge that affects the Wnt/β-catenin signaling pathway and may also affect other signaling pathways. Wang ST et al found that circ-ITCH acts as a sponge of miR-214 and miR-17, increasing the expression of its ITCH linear isoform, thereby inactivating the Wnt/β-catenin signal, thereby showed that circ-ITCH is a tumor suppressor, a promising prognostic biomarker in triple-negative breast cancer (TNBC) and that its restoration could well be a successful strategy in TNBC;42 Wang MN et al revealed a novel signaling pathway of circ-ITCH/miR-22-3p/CBL/β-catenin involved in papillary thyroid cancer (PTC) development and progression;43 Li F et al suggested that cir-ITCH is a tumor-suppressor gene in glioma and may serve as a promising prognostic biomarker for glioma patients by sponging miR-214 and regulating ITCH-Wnt/β-catenin pathway;44 Yang CD et al concluded that circ-ITCH acts as a tumor suppressor by a novel circ-ITCH/miR-17, miR-224/p21, PTEN axis, which may provide a potential biomarker and therapeutic target for the management of bladder cancer.45 Hu JH et al provided a novel tumor suppressive role regarding circ-ITCH function in the malignant progression of ovarian cancer through targeting miR-145/RASA1 signaling.46
SNF2 Histone Linker PHD RING Helicase (SHPRH)
The coding gene for circ-SHPRH is located at 6q24.3 and contains a total of 37 exons. The SHPRH gene is present in the ovary (RPKM 1.7), thyroid (RPKM 1.5) and 25 other tissues. SHPRH is a ubiquitously expressed protein containing motifs of several DNA repair proteins, transcription factors and helicases. SHPRH is a functional homolog of Rad5.78
Recent studies indicated that circ-SHPRH could be seen as biomarkers and therapeutic targets in many cancers. Zhang ML et al discovered that a novel protein SHPRH-146aa generated from overlapping genetic codes of circ-SHPRH is a tumor suppressor in human glioblastoma;47 Qin ML et al indicated that hsa_circ_0001649 (circ-SHPRH) might serve as a novel potential biomarker for HCC and may function in tumorigenesis and metastasis of HCC;48 Xu Y et al suggested that hsa_circ_0001649 might be a rational CCA-related therapeutic target based on its overexpression caused tumor suppressive effects via inhibiting cell proliferation, migration and invasion, inducing cell apoptosis in KMBC and Huh-28 cells;49 Xing LC et al reported that circ-SHPRH might be a potentially useful prognostic biomarker and therapeutic target for Retinoblastoma via affect AKT/mTOR signaling pathway.50
Protein Kinase C Iota (PRKCI)
The coding gene for circPRKCI is located at 3q26.2 and contains 18 exons. The gene PRKCI is ubiquitously expressed in the stomach (RPKM 19.1), thyroid (RPKM 17.9) and other 24 tissues. This gene encodes serine/threonine protein kinase as a member of the protein kinase C (PKC) family. The PKC family contains at least 8 members that are differentially expressed and involved in various cellular processes. This kinase can be recruited into vesicular tubular clusters (VTC) through direct interaction with small gtpase RAB2, which plays a role in tubular kinetics by phosphorylating glyceraldehyde 3-phosphodehydrogenase (GAPD/GAPDH) and microRNA in the early secretory pathway.
Shi NM et al and Xia WJ et al revealed that circ-PRKCI plays an important role in esophageal squamous cell carcinoma (ESCC) by affecting miR-3680-3p thus regulated AKT3 expression and that provide new insights into the pathogenesis of ESCC;21,52 Wang HH et al concluded that circ_0067934(circ-PRKCI) could improve the development of thyroid carcinoma by promoting epithelial-mesenchymal-transition (EMT) and PI3K/AKT signaling pathways;53 Hu CJ et al revealed that circ-PRKCI promotes cervical cancer progression via miR-545/EIF3C axis;54 Zou Q et al and Wang J et al indicated that circ-PRKCI may be a predictive marker for the prognosis of non-small cell lung cancer (NSCLC) and a target for the treatment of the disease;55,56 Zhu Q et al summarized that the circ-PRKCI/miR-1324/FZD5/β-catenin signaling axis might serve as a promising therapeutic target for HCC intervention.57
Adaptor Related Protein Complex 4 Subunit Epsilon 1 (AP4E1)
The coding gene for circAP4E1 is located at 15q21.2 and contains a total of 23 exons. The gene AP4E1 is present in lymph nodes (RPKM 2.9), testes (RPKM 2.9) and 25 other tissues. The gene encodes members of the large subunit protein family of the linker complex. These proteins are components of the cohesive protein complex and play an important role in the secretory and endocytic pathways by mediating vesicular formation and the sorting of integrative membrane proteins.
Liu W et al illustrated a novel hsa_circRNA_103809 (circ-AP4E1/miR-4302/ZNF121/MYC regulatory signaling pathway promotes lung cancer progression;58 Bian LJ et al and Zhang PL et al indicated that circ-AP4E1 may be a potential novel gene target for the diagnosis and treatment of colorectal cancer (CRC) by participating in the regulation of biological functions through the miR-532-3P/FOXO4 axis;59,60 Liu WH et al indicated that circRNAs may serve as biomarkers of osteosarcoma diagnosis and treatment;61 Li X et al and Cai HJ et al concluded that circ-AP4E1 may serve as a potential biomarker and novel therapeutic target of HCC by binding to miR-620 and inhibiting the tumourigenicity of HCC,62 and facilitating HCC malignant progression by regulating miR-490-5p/SOX2 signaling pathway.63
DEAD-Box Helicase 42 (DDX42)
The coding gene for circ-DDX42 is located at 17q23.3 and contains a total of 20 exons. The gene DDX42 is ubiquitously expressed in testis (RPKM 27.4), ovary (RPKM 25.5) and 25 other tissues. The gene encodes a member of the Asp-Glu-Ala-Asp (DEAD) casein family. Members of this protein family are presumed RNA helicases and are involved in cellular processes involved in RNA secondary structural changes.
Song LL et al suggested an oncogenic role for hsa_circ_0007534 (circ-DDX42) in breast cancer by acting as a miR-593 sponge to promote MUC19 expression;64 Zhang R et al concluded that circ-DDX42 plays a crucial role in the initiation and progression of CRC and may be a potential therapeutic target of CRC;65 Li BQ et al summarized that circ-DDX42 may be a rational predictive marker and therapeutic target for osteosarcoma by affecting AKT/GSK-3β signaling pathway;66 Li GF et al suggested that circ-DDX42/miR-761/ZIC5 axis might be a target for glioma treatment;67 Hao LG et al illuminated a novel circRNA (circ-DDX42) that confers an oncogenic function in pancreatic ductal adenocarcinoma.(PDAC) by sponging miR-625 and miR-892b.68
Ubiquitin Associated Protein 2 (UBAP2)
The coding gene for circUBAP2 is located at 9q13.3 and contains a total of 30 exons. The gene UBAP2 is widely expressed in testis (RPKM 18.9), adrenal gland (RPKM 7.6) and other 25 tissues. The protein encoded by this gene contains a UBA (ubiquitin-associated) domain, which is characteristic of proteins that play a role in the ubiquitination pathway. This gene may be expressed in adrenal and lymphoid tissues.
Zhou JX et al concluded that hsa_circ_0008344(circ-UBAP2) is upregulated in glioblastoma and may contribute to the progression of this malignancy;69 Zhang H et al suggested that circ-UBAP2 sponging miR-143 to promote osteosarcoma progression and implicate its potential in prognosis prediction and cancer therapy;70 Wang ST et al revealed that circ-UBAP2 plays a vital regulatory role in TNBC via the miR-661/MTA1 axis and may serve as a promising therapeutic target for TNBC patients;71 Yin YJ et al concluded that circ-UBAP2 exerts an important role in the proliferation and invasion of human lung cancer. Silencing of circUBAP2 might be a novel target for molecular targeted therapy of patients with lung cancer.72
Conclusion And Perspective
In the past, researchers generally believed that circRNAs were splicing errors and that the intermediates were detached from the intron sleeve, so circRNAs were not considered to have important functions. In recent years, with the rapid development of high-throughput sequencing, it has been found that circRNAs play an indispensable role in the occurrence and development of many diseases through biological functions such as miRNA sponge, protein binding molecules and transcriptional regulators. Due to the large number of circRNAs in blood and tissue samples, many scholars have pointed out that circRNAs are biomarkers for cancer screening, prognosis assessment, clinical diagnosis and cancer treatment goals. In the future, circRNAs have great potential in these areas.
However, studies on circRNAs in cancer recurrence and metastasis are still incomplete and require further exploration. Based on this state, rapid development of multiple displacement amplification (MDA), multiple annealing and cycle-based amplification cycles, MALBAC, single-cell sequencing and next-generation sequencing can greatly improve sequencing efficiency during the development phase. Accuracy helps identify new circRNAs and also helps researchers perform small sequence variation detection, individual cytogenetic differences, gene rearrangements, etc., which can help explain cancer recurrence, metastasis, and progression. At the same time, it opens up new ideas for studying various complex diseases and physiological processes from the single nucleotide level. Although many functions of circRNAs have been discovered and the number of circRNAs has some function, there are still thousands of circRNAs with unknown functions. Further studies of the biogenesis of circRNAs may be needed to discover more functions of circRNAs. Due to its classicity and high research maturity, most existing circRNA studies focus on the mechanism by which miRNAs adsorb miRNAs. In fact, although research in this area is not thorough enough, the ability of circRNAs to work by binding to DNA or proteins is also promising. In addition, circRNAs are different from linear RNA cleavage and deserves further study. For example, circus splicing abnormalities are a very good research direction under physiological and pathological conditions.
The ultimate goal of medical research is to provide services for clinical diagnosis and treatment. How to apply the research results of circRNAs to clinical practice requires more relevant research to support it, such as the design of molecularly targeted drugs for specific regions of circRNAs. Drug delivery and the discovery of circRNAs involved in the development of most cancers. The work of this review is to try to discover circRNAs that are involved in a variety of cancers. We hope that with the development of biotechnology and basic research, we will be able to discover more circRNAs and more physiological and pathological circRNA functions, so that circRNAs can better serve clinical diagnosis and treatment.
Acknowledgments
This project was supported by Fundamental Research Funds for the Central Universities (2242018K40168) for Dr. Ziyi Chen and a grant from the Medical Science and Technology Development Foundation (ZKX18027) for Hanjin Wang.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852–3856. doi:10.1073/pnas.73.11.3852
2. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. doi:10.1038/323558a0
3. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi:10.1038/nrm.2015.32
4. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi:10.1080/15476286.2015.1020271
5. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi:10.1038/323558a0
6. Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. Fabes J. 1993;7:155–160.
7. Xu ZJ, Yan YL, Zeng SS, et al. Circular RNAs: clinical relevance in cancer. Oncotarget. 2017;9(1):1444–1460. doi:10.18632/oncotarget.22846
8. Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21(2):172–179.
9. Zhang XO, Dong R, Zhang Y, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26(9):1277–1287.
10. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
11. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi:10.1371/journal.pgen.1003777
12. Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi:10.1016/j.molcel.2013.08.017
13. Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. Embo J. 2011;30(21):4414–4422. doi:10.1038/emboj.2011.359
14. Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M. Circular RNA: an emerging key player in RNA world. Brief Bioinform. 2017;18(4):547–557. doi:10.1093/bib/bbw045
15. Wu Q, Li P, Wu M, Liu Q. Deregulation of circular RNAs in cancer from the perspectives of aberrant biogenesis, transport and removal. Front Genet. 2019;1(10):16. doi:10.3389/fgene.2019.00016
16. Zhang ZK, Xie Q, He DM, et al. Circular RNA: new star, new hope in cancer. BMC Cancer. 2018;18(1):834. doi:10.1186/s12885-018-4242-8
17. Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA. 2017;8:2. doi:10.1002/wrna.1386
18. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi:10.1371/journal.pone.0030733
19. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi:10.1016/j.cell.2004.12.035
20. Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11:e0158347. doi:10.1371/journal.pone.0158347
21. Xu B, Yang TY, Wang Z, Zhang Y, Liu SY, Shen MQ. CircRNA CDR1as/miR-7 signals promote tumor growth of osteosarcoma with a potential therapeutic and diagnostic value. Cancer Manag Res. 2018;10:4871–4880. doi:10.2147/CMAR.S178213
22. Xu LL, Zhang M, Zheng XB, Yi PS, Lan C, Xu MQ. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2016;143:17–27. doi:10.1007/s00432-016-2256-7
23. Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi:10.1038/nsmb.2959
24. Zhang Y, Zhang XO, Chen T, et al. Circular intronic long non coding RNAs. Mol Cell. 2013;51:792–806. doi:10.1016/j.molcel.2013.04.023
25. Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15(8):995–1005. doi:10.1080/15476286.2018.1486659
26. Zang JK, Lu D, Xu AD. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neuro Res. Epub 2018 Dec 21.
27. Janas T, Janas MM, Sapon K, Janas T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015;589(13):1391–1398. doi:10.1016/j.febslet.2015.04.036/
28. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13(1):34–42. doi:10.1080/15476286.2015.1119365
29. Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and characterizing circRNA-protein interaction. Theranostics. 2017;7(17):4183–4191. doi:10.7150/thno.21299
30. Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. Embo J. 2013;32(7):923–925. doi:10.1038/emboj.2013.53
31. Zhang Y, Xue W, Li X, et al. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15(3):611–624. doi:10.1016/j.celrep.2016.03.058
32. Jiang XM, Li ZL, Li JL, et al. A novel prognostic biomarker for cholangiocarcinoma: circRNA Cdr1as. Eur Rev Med Pharmacol Sci. 2017;22:365–371.
33. Xu LL, Zhang M, Zheng XB, Yi PS, Lan C, Xu MQ. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2016;143:17–27. doi:10.1007/s00432-016-2256-7
34. Xu HY, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi:10.1038/srep12453
35. Li XB, Zheng YF, Zheng Y, et al. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther. 2018;9:232. doi:10.1186/s13287-018-0976-0
36. Yao WX, Li Y, Han L, et al. The CDR1as/miR-7/TGFBR2 axis modulates EMT in silica-induced pulmonary fibrosis. Toxicol Sci. 2018;166(2):465–478. doi:10.1093/toxsci/kfy221
37. Li P, Yang X, Yuan WB, et al. CircRNA-Cdr1as exerts anti-oncogenic functions in bladder cancer by sponging MicroRNA-135a. Cell Physiol Biochem. 2018;46:1606–1616. doi:10.1159/000489208
38. Shuai MX, Hong JW, Huang DH, Zhang X, Tian YQ. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol Lett. 2018;16:6495–6501. doi:10.3892/ol.2018.9471
39. Jin PC, Huang YN, Zhu PL, Zou Y, Shao TT, Wang OY. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem Biophys Res Commun. 2018;503:1570–1574. doi:10.1016/j.bbrc.2018.07.081
40. Ding K, Liao YN, Chen YT, Gong DH, Zhao X, Ji W. Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem Biophys Res Commun. 2018;503(2018):863–869. doi:10.1016/j.bbrc.2018.06.088
41. Ma XL, Zhu KP, Zhang CL. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9(10):1856–1862. doi:10.7150/jca.24619
42. Wang ST, Liu LB, Li XM, et al. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/-catenin pathway. NEOPLASMA. 2018. doi:10.4149/neo_2018_180710N460
43. Wang MN, Chen B, Ru ZX, Cong L. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/b-catenin pathway. Biochem Biophys Res Commun. 2018;504:283–288. doi:10.1016/j.bbrc.2018.08.175
44. Li F, Ma K, Sun MH, Shi S. Identification of the tumor-suppressive function of circular RNA ITCH in glioma cells through sponging miR-214 and promoting linear ITCH expression. Am J Transl Res. 2018;10(5):1373–1386.
45. Yang CD, Yuan WB, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19. doi:10.1186/s12943-018-0771-7
46. Hu JH, Wang L, Chen JM, et al. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem Biophys Res Commun. 2018;505:222–228. doi:10.1016/j.bbrc.2018.09.060
47. Zhang ML, Huang NN, Yang XS, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi:10.1038/s41388-017-0019-9
48. Qin ML, Liu G, Huo XS, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomarkers. 2016;16(2016):161–169. doi:10.3233/CBM-150552
49. Xu Y, Yao Y, Zhong XY, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496:455–461. doi:10.1016/j.bbrc.2018.01.077
50. Xing LC, Zhang LM, Feng YL, Cui Z, Ding L. Downregulation of circular RNA hsa_circ_0001649 indicates poor prognosis for retinoblastoma and regulates cell proliferation and apoptosis via AKT/mTOR signaling pathway. Biomed Pharmacother. 2018;105(2018):326–333. doi:10.1016/j.biopha.2018.05.141
51. Shi NM, Shan B, Gu B, Song YQ, Chu HJ, Qian L. Circular RNA circ-PRKCI functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-3680-3p in esophageal squamous cell carcinoma. J Cell Biochem. 2018;2019:1–10.
52. Xia WJ, Qiu MT, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi:10.1038/srep35576
53. Wang HH, Yan XG, Zhang HJ, Zhan XR. CircRNA circ_0067934 overexpression correlates with poor prognosis and promotes thyroid carcinoma progression. Med Sci Monitor. 2019;25:1342–1349. doi:10.12659/MSM.913463
54. Hu CJ, Wang Y, Li A, Zhang J, Xue FF, Zhu L. Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR-545/EIF3C axis. J Cell Physiol. 2019;234(6):9225–9232.
55. Zou QG, Wang TJ, Li B, et al. Overexpression of circ-0067934 is associated with increased cellular proliferation and the prognosis of non-small cell lung cancer. Oncol Lett. 2018;16:5551–5556. doi:10.3892/ol.2018.9357
56. Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci. 2018;22:3053–3060. doi:10.26355/eurrev_201805_15063
57. Zhu Q, Lu GY, Luo ZH, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochem Biophys Res Commun. 2018;02:119.
58. Liu W, Ma WM, Yuan Y, Zhang YW, Sun SY. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500:846–851. doi:10.1016/j.bbrc.2018.04.172
59. Bian LJ, Zhi XF, Ma LL, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532e3p/FOXO4 axis. Biochem Biophys Res Commun. 2018;505:346–352. doi:10.1016/j.bbrc.2018.09.073
60. Zhang PL, Zuo ZG, Shang WJ, et al. Identification of differentially expressed circular RNAs in human colorectal cancer. Tumor Biol. 2017;(2017):1–10.
61. Liu WH, Zhang JJ, Zou CY, et al. Microarray expression profile and functional analysis of circular RNAs in osteosarcoma. Cell Physiol Biochem. 2017;43:969–985. doi:10.1159/000481650
62. Li X, Shen M. Circular RNA hsa_circ_103809 suppresses hepatocellular carcinoma proliferation and invasion by sponging miR-620. Eur Rev Med Pharmacol Sci. 2019;23:555–566. doi:10.26355/eurrev_201902_16868
63. Cai HJ, Hu BR, Ji L, Ruan XJ, Zheng ZH. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway. Am J Transl Res. 2018;10(6):1690–1702.
64. Song LL, Xiao Y. Downregulation of hsa_circ_0007534 suppresses breast cancer cell proliferation and invasion by targeting miR-593/MUC19 signal pathway. Biochem Biophys Res Commun. 2018;503:2603–2610. doi:10.1016/j.bbrc.2018.08.007
65. Zhang R, Xu J, Zhao J, Wang X. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci. 2018;22:118–126. doi:10.26355/eurrev_201801_14108
66. Li BQ, Li XG. Overexpression of hsa_circ_0007534 predicts unfavorable prognosis for osteosarcoma and regulates cell growth and apoptosis by affecting AKT/GSK-3β signaling pathway. Biomed Pharmacother. 2018;107:860–866. doi:10.1016/j.biopha.2018.08.086
67. Li GF, Li L, Yao ZQ, Zhuang SJ. Hsa_circ_0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells. Biochem Biophys Res Commun. 2018;499:765–771. doi:10.1016/j.bbrc.2018.03.219
68. Hao LG, Rong W, Bai LJ, et al. Upregulated circular RNA circ_0007534 indicates an unfavorable prognosis in pancreatic ductal adenocarcinoma and regulates cell proliferation, apoptosis, and invasion by sponging miR-625 and miR‐892b. J Cell Biochem. 2019;120(3):3780–3789.
69. Zhou JX, Wang HX, Chu JS, et al. Circular RNA hsa_circ_0008344 regulates glioblastoma cell proliferation, migration, invasion, and apoptosis. J Clin Lab Anal. 2018;e22454. doi:10.1002/jcla.22454
70. Zhang H, Wang GC, Ding C, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8(37):61687–61697. doi:10.18632/oncotarget.18671
71. Wang ST, Li Q, Wang YF, et al. Upregulation of circ-UBAP2 predicts poor prognosis and promotes triple-negative breast cancer progression through the miR-661/MTA1 pathway. Biochem Biophys Res Commun. 2018;505:996–1002. doi:10.1016/j.bbrc.2018.10.026
72. Yin YJ, Gao H, Guo J, Gao Y. Effect of circular RNA UBAP2 silencing on proliferation and invasion of human lung cancer A549 cells and its mechanism. Chin J Lung Cancer. 2017;20:12.
73. Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73.
74. Qi H, Sun Y, Jiang Y, Li X. Upregulation of circular RNA circ_0000502 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-1238. J Cell Biochem. 2019;120(5):8475-8482. doi:10.1002/jcb.28134
75. Xu Y, Yao Y, Liu Y, et al. Elevation of circular RNA circ_0005230 facilitates cell growth and metastasis via sponging miR-1238 and miR-1299 in cholangiocarcinoma. Aging (Albany NY). 2019;Apr. doi:10.18632/aging.101872
76. Xu LL, Feng XF, Hao XY, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38(1):98.
77. Chen L, Zhou H, Guan Z. CircRNA_000543 knockdown sensitizes nasopharyngeal carcinoma to irradiation by targeting miR-9/platelet-derived growth factor receptor B axis. Biochem Biophys Res Commun. 2019;512(4):786–792. doi:10.1016/j.bbrc.2019.03.126
78. Unk I, Hajdú I, Fátyol K, et al. Human SHPRH is a ubiquitin ligase for Mms2–ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2006;103(48):18107–18112.
79. Shi F, Shi ZH, Zhao YD, Tian JW. CircRNA hsa-circ-0014359 promotes glioma progression by regulating miR-153/PI3K signaling. Biochem Biophys Res Commun. 2019;510(4):614–620.
80. Li X, Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J Cell Physiol. 2019;234(8):13807–13819. doi:10.1002/jcp.28061
81. Yang M, Li G, Fan L, Zhang G, Xu J, Zhang J. Circular RNA circ_0034642 elevates BATF3 expression and promotes cell proliferation and invasion through miR-1205 in glioma. Biochem Biophys Res Commun. 2019;508(3):980–985.
82. Lei B, Huang Y, Zhou Z, et al. Circular RNA hsa_circ_0076248 promotes oncogenesis of glioma by sponging miR-181a to modulate SIRT1 expression. J Cell Biochem. 2019;120(4):6698–6708.
83. Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18(1):71.
84. Ding L, Zhao Y, Dang S, et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18(1):45.
85. Lu J, Zhang PY, Li P, et al. Circular RNA hsa_circ_0001368 suppresses the progression of gastric cancer by regulating miR-6506-5p/FOXO3 axis. Biochem Biophys Res Commun. 2019;512(1):29–33.
86. Rong D, Lu C, Zhang B, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. 2019;18(1):25.
87. Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20.
88. Zhang H, Wang X, Huang H, Wang Y, Zhang F, Wang S. Hsa_circ_0067997 promotes the progression of gastric cancer by inhibition of miR-515-5p and activation of X chromosome-linked inhibitor of apoptosis (XIAP). Artif Cells Nanomed Biotechnol. 2019;47(1):308–318.
89. Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10(3):175.
90. Tang H, Huang X, Wang J, et al. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol Cancer. 2019;18(1):23.
91. Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18(1):4.
92. Liu W, Zhao J, Jin M, Zhou M. circRAPGEF5 contributes to papillary thyroid proliferation and metastatis by regulation miR-198/FGFR1. Mol Ther Nucleic Acids. 2019;14:609–616.
93. Pan Y, Xu T, Liu Y, Li W, Zhang W. Upregulated circular RNA circ_0025033 promotes papillary thyroid cancer cell proliferation and invasion via sponging miR-1231 and miR-1304. Biochem Biophys Res Commun. 2019;510(2):334–338.
94. Cao W, Zhao Y, Wang L, Huang X. Circ0001429 regulates progression of bladder cancer through binding miR-205-5p and promoting VEGFA expression. Cancer Biomark. 2019. doi:10.3233/CBM-182380
95. Lin G, Sheng H, Xie H, et al. circLPAR1 is a novel biomarker of prognosis for muscle-invasive bladder cancer with invasion and metastasis by miR-762. Oncol Lett. 2019;17(3):3537–3547.
96. Su H, Tao T, Yang Z, et al. Circular RNA cTFRC acts as the sponge of MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer. 2019;18(1):27.
97. Li Y, Wan B, Liu L, Zhou L, Zeng Q. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR-221 and inhibiting epithelial-to-mesenchymal transition. Biochem Biophys Res Commun. 2019;508(4):991–996.
98. Song H, Xu D, Shi P, et al. Upregulated circ RNA hsa_circ_0000337 promotes cell proliferation, migration, and invasion of esophageal squamous cell carcinoma. Cancer Manag Res. 2019;11:1997–2006.
99. Mao Y, Zhang L, Li Y. circEIF4G2 modulates the malignant features of cervical cancer via the miR-218/HOXA1 pathway. Mol Med Rep. 2019;19(5):3714–3722.
100. Song T, Xu A, Zhang Z, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol. 2019. doi:10.1002/jcp.28128
101. Cai H, Zhang P, Xu M, Yan L, Liu N, Wu X. Circular RNA hsa_circ_0000263 participates in cervical cancer development by regulating target gene of miR-150-5p. J Cell Physiol. 2019;234(7):11391–11400. doi:10.1002/jcp.27796
102. Liu J, Wang D, Long Z, Liu J, Li W. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p/519-5p family and modulating the expression of CBX8. Cell Physiol Biochem. 2018;48(1):173–184. doi:10.1159/000491716
103. Liu G, Shi H, Deng L, et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;
104. An J, Shi H, Zhang N, Song S. Elevation of circular RNA circ_0003645 forecasts unfavorable prognosis and facilitates cell progression via miR-1179/TMEM14A pathway in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;511(4):921–925. doi:10.1016/j.bbrc.2019.03.011
105. Min L, Wang H, Zeng Y. CircRNA_104916 regulates migration, apoptosis and epithelial-mesenchymal transition in colon cancer cells. Front Biosci (Landmark Ed). 2019;24:819–832.
106. Li XN, Wang ZJ, Ye CX, Zhao BC, Huang XX, Yang L. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacother. 2019;112:108611. doi:10.1016/j.biopha.2019.108611
107. Liu L, Liu FB, Huang M, et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. 2019;pii:
108. Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochem Biophys Res Commun. 2019;509(1):138–142. doi:10.1016/j.bbrc.2018.12.088
109. Qu S, Hao X, Song W, et al. Circular RNA circRHOT1 is upregulated and promotes cell proliferation and invasion in pancreatic cancer. Epigenomics. 2019;11(1):53–63. doi:10.2217/epi-2018-0051
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.