Back to Journals » OncoTargets and Therapy » Volume 12
SPOP Regulates The Biological Mechanism Of Ovarian Cancer Cells Through The Hh Signaling Pathway
Authors Li Y, Yu Q , Li R, Luo J, Yuan D, Song J, Sun Y , Long T, Yang Z
Received 26 May 2019
Accepted for publication 24 September 2019
Published 6 November 2019 Volume 2019:12 Pages 9239—9248
DOI https://doi.org/10.2147/OTT.S215940
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Yanxi Li,1 Qiubo Yu,2 Ruohan Li,1 Jing Luo,1 Dong Yuan,1 Jiao Song,1 Yixuan Sun,1 Tengfei Long,1 Zhu Yang1
1Department of Gynecology, The Second Affiliated Hospital of Chongqing Medical University, Chongqing 400010, People’s Republic of China; 2Molecular Medical Testing Center, Chongqing Medical University, Chongqing 400016, People’s Republic of China
Correspondence: Zhu Yang
Department of Gynecology, The Second Affiliated Hospital of Chongqing Medical University, 76 Linjiang Road, Chongqing 400010, People’s Republic of China
Tel/Fax +86 23 6848 6646
Email [email protected]
Background: Ovarian cancer is characterized by high metastatic potential and high mortality. More than 80% of primary ovarian malignancies are epithelial ovarian cancers. There is increasing evidence that Speckle-type POZ protein (SPOP) is highly correlated with the development of various types of cancer. However, the effects of SPOP on epithelial ovarian cancer and the associated molecular mechanisms remain unclear.
Materials and methods: We compared SPOP expression between epithelial ovarian cancer tissues and normal ovarian tissues by using immunohistochemical staining. To determine the role of SPOP in epithelial ovarian cancer cells, we overexpressed or knocked down SPOP in the epithelial ovarian cancer cell line OVCAR-3 using lentiviral vectors.
Results: Our results from the present study indicated that SPOP expression was significantly downregulated in human epithelial ovarian cancer and was associated with the FIGO stage and the histopathologic grading of the tumor. The overexpression and knockdown experiments revealed that SPOP inhibited proliferation while promoting apoptosis in ovarian cancer cells. Inhibition of SPOP mis-activated the Hedgehog (Hh) signaling pathway, thereby inhibiting apoptosis in ovarian cancer cells.
Conclusion: SPOP suppresses proliferation and promotes apoptosis in human ovarian cancer cells by inhibiting the Hh signaling pathway, offering the possibility of new approaches for the treatment of ovarian cancer.
Keywords: ovarian cancer, SPOP, Hedgehog signaling, apoptosis, proliferation
Introduction
Ovarian cancer is one of the most fatal gynecological tumors. More than 80% of primary ovarian malignancies are epithelial ovarian cancers. It is estimated that there are approximately 239,000 new cases of ovarian cancer and 152,000 related deaths worldwide each year.1 Ovarian cancer is often referred to as a “silent killer” because patients tend to be in an advanced stage with poor prognosis by the time the ovarian cancer is diagnosed.2 The occurrence of ovarian cancer is closely related to chromosomal abnormalities, oncogene activation, and inactivation of tumor suppressor genes. Although considerable progress has been made in understanding the molecular mechanisms underlying ovarian cancer progression, the prognosis of patients with advanced ovarian cancer remains poor. Therefore, it is necessary to further explore the molecular mechanisms, whereby new therapeutic interventions can be developed.
The human SPOP gene is located on chromosome 17q21.33. Heterozygous deletion (LOH) of the SPOP gene locus has been observed in a high percentage of the human breast cancer cases.3 Previous studies have found high-frequency mutations in the SPOP gene in prostate and endometrial cancer.4,5 SPOP is a CUL3-based E3 ubiquitin ligase adaptor protein,6 containing the following four domains: an N-terminal MATH domain responsible for substrate recruitment, a BTB domain for the interaction with cullin3, a C-terminal nuclear localization sequence, and a 3-box domain that facilitates binding to cullin3.7 SPOP promotes ubiquitination and proteasomal degradation of its substrates. Multiple SPOP substrates have recently been identified, including AR, steroid receptor coactivator SRC-3, ERG, ERα, and PR.8–13 They all have been found to contain a common SBC motif. SPOP has been identified as a key tumor suppressor that promotes degradation of several oncogenic proteins in various cancers, including prostate, gastric, endometrial, liver, glioma, colorectal, breast, and osteosarcoma cancers.12–20 However, in clear cell renal cell carcinoma, SPOP has been reported to be upregulated and accumulated in the cytoplasm under hypoxic conditions, supporting the progression of the cancer by promoting ubiquitination and degradation of PTEN, DUSP7, and DAXX.21 The differential effect of SPOP in renal clear cell carcinoma and other cancers may be due to differences in the subcellular localization of SPOP in different cancers. In addition, we have previously identified high-frequency LOH in the SPOP locus in ovarian cancer tissues by fluorescence in situ hybridization (FISH) and suggested that SPOP may play a role in ovarian cancer.22 However, such a role and the related mechanisms remain unclear.
The Hedgehog (Hh) signaling pathway is a morphogenetic pathway critical for the formation and growth of various tissues during embryonic development.23,24 There is growing evidence that the Hh pathway is mis-activated in almost all tumors, including ovarian cancer.25–29 The Hh signaling in mammals encompasses three Hh ligands (Sonic hedgehog-Shh, Indian hedgehog-Ihh, and Desert hedgehog-Dhh), two Patched receptors (PTCH1 and PTCH2), the signal transducer protein Smoothened (SMO), and three transcription factors (Gli1, Gli2, and Gli3), for which suppressor of fused (SuFu) functions as a negative regulator, preventing activation of Hh target genes.30 In the absence of an Hh ligand, Smo activation is inhibited by the 12-transmembrane domain protein PTCH located in the cell membrane. Binding of an Hh ligand to PTCH releases this inhibition, allowing Smo to activate the Gli family transcription factors. These transcription factors then translocate to the nucleus and activate transcription of Hh target genes, which promote cell survival, proliferation, and differentiation.31 The Drosophila homolog of SPOP, called HIB or rdx, inhibits the Hh signaling by mediating degradation of the sole Gli transcription factor Ci in the fly.32,33 Interestingly, SPOP has been found to interact with Gli2 in gastric cancer and colorectal tumors through post-transcriptional modification and to downregulate Gli2 in renal cancer, thereby exerting a tumor-suppressive effect.16,19
In this study, we evaluated the expression of SPOP in ovarian cancer and normal tissues. We then analyzed the possible correlations between the clinicopathological features and SPOP expression. Based on the clinical findings, we performed in vitro experiments and studied the effect of up- or down-regulation of SPOP on ovarian cancer cell proliferation and apoptosis. We observed that SPOP had a pro-apoptotic effect in ovarian cancer cells by modulating Hh signaling.
Materials And Methods
Reagents And Antibodies
The Cell-light™ EdU Apollo567 In Vitro Kit was purchased from RiboBio (C10310–1; RiboBio,Guangzhou,China. The Gli inhibitor GANT61 was purchased from MedchemExpress (HY-13901; MedchemExpress, Monmouth Junction, NJ, USA). Dimethyl sulfoxide (DMSO) was purchased from Sigma (Sigma, Milwaukee, WI, USA). Antibodies against the following proteins were used for Western blotting at the dilutions indicated in parentheses: Hedgehog signaling antibody sampler kit (8358; 1:1000), caspase3 (9668; 1:1000), GAPDH (5174; 1:1000), and Bax (2772; 1:1000) were purchased from Cell Signaling Technology (Cell Signaling Technology, Beverly, MA, USA); SPOP (ab168619; 1:1000) was purchased from Abcam (Abcam, Cambridge, MA, USA); Bcl2 (bs-0032R; 1:500) and PCNA (bs-0754R; 1:500) were purchased from Bioss (Bioss, Beijing, China); and GLI2 (WL02691; 1:500) was purchased from Wanleibio (Wanleibio, Shenyang, China). SPOP (16750-1-AP; 1:200) antibody for immunohistochemistry was purchased from Proteintech (Proteintech, Wuhan, China).
Sample Collection
Paraffin sections of human ovarian cancer and healthy tissue samples from 55 and 30 individuals, respectively, were obtained from the Chongqing Medical University Pathology Testing Center. Ovarian epithelial cancer tissue and normal ovarian epithelial tissue constituted the experimental and control groups, respectively. Of the 55 carcinoma cases, 7 cases were diagnosed at stage I, 10 cases at stage II, 29 cases at stage III, and 9 cases at stage IV according to the International Federation of Gynecology and Obstetrics (FIGO) classification. None of the patients had any tumor other than primary ovarian epithelial cancer, and none had received chemotherapy or radiotherapy before the surgery. The control samples originated from individuals undergoing total abdominal hysterectomy for uterine fibroids, and none of these individuals had any malignant tumors in other organs before the operation. The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the Second Affiliated Hospital of Chongqing Medical University.
Immunohistochemical Staining
The tissue sections were de-paraffinized with xylene for 30 min and rehydrated with decreasing concentrations of ethanol. Next, they were immersed in citrate buffer, heated in a microwave oven for 5 min, and then maintained at medium heat for 10 min for antigen retrieval. Immunohistochemical staining was performed according to the instructions in the polymer detection system (PV-9000; Zsbio, Beijing, China). The sections were treated with the anti-SPOP antibody overnight at 4 °C, and the bound antibodies were visualized with 3,3ʹ-diaminobenzidine (DAB; Zsbio). The experimental and control samples were examined with a microscope under the same exposure conditions (OLYMPUS, Tokyo, Japan). For each sample, a comprehensive scoring was performed based on the intensity of SPOP protein staining (no stain = 0; light yellow = 1; yellow-brown = 2; brown = 3) and the percentage of stained cells (No staining = 0, 1–24% = 1, 25–49% = 2, 50–74% = 3, 75–100% = 4). The final immunohistochemical score was determined by multiplying the intensity score by the percentage score, ranging from 0 (minimum) to 12 (maximum).
Cell Culture And Transfection
OVCAR-3 cells were cultured in RPMI-1640 (Gibco, NY, USA) containing 10% fetal bovine serum (FBS; Cellmax, Lanzhou, China) and 1% antibiotic mixture (Beyotime, Shanghai, China) at 37 °C in a humidified incubator with 5% CO2. A total of 3 × 105 cells were infected with a lentivirus (Sunbio Medical Biotechnology, Shanghai, China) carrying the SPOP gene (ov-SPOP), SPOP shRNA (sh-SPOP), or negative control (ov-NC or sh-NC) following the manufacturer’s instructions. The infected cells were selected with 72 h of incubation with 10-µg/mL puromycin (P8230; Solarbio, Beijing, China).
Western Blot Analysis
The cells were washed with cold PBS and lysed in RIPA buffer (P0013B, Beyotime) containing 1% PMSF (ST506, Beyotime). The cell lysates were centrifuged at 12,000 × g for 15 min to remove the debris, and the supernatants were kept for further analysis. The samples were mixed with 25% SDS-PAGE Sample Loading Buffer (P0015; Beyotime) and incubated at 100 °C for 3–5 min. They were then run on an 8–12% SDS-PAG and transferred onto a polyvinylidene fluoride membrane. Subsequently, the membrane was blocked with 2 h of incubation with 5% BSA (SW3015, Solarbio) at room temperature and incubated with the antibodies overnight at 4 °C. Horseradish peroxidase (HRP)-conjugated secondary antibodies were used to detect the primary antibodies. The ECL detection system (WBKLS0100; Millipore, Billerica, MA, USA) was used to visualize the protein bands. The expression values of the proteins analyzed were normalized to the expression of GAPDH. All the experiments were repeated at least three times and yielded consistent results.
Colony Formation Assay
Five hundred cells were seeded in each well of a 6-well plate and incubated in RPMI-1640 with 10% FBS at 37 °C. Two weeks later, the cells were fixed with 5% polyacetal and stained with 0.1% crystal violet. Colonies with > 50 cells were counted. The experiments were performed in triplicate.
qRT-PCR
Total RNA (1 μg) was used to prepare cDNA by reverse transcription using a PrimeScript™ RT Reagent Kit (RR037A; TaKaRa, Tokyo, Japan) according to the manufacturer’s instructions. qRT-PCR was performed using SYBR Premix Ex TaqTM II (RR820A; TaKaRa). The data were analyzed using the comparative ∆∆CT method. The target gene mRNA level was normalized to that of GAPDH. The following primers were used for the PCR: SPOP (forward, 5′-GCCCTCTGCAGTAACCTGTC-3′; reverse, 5′-GTCTCCAAGACATCCGAAGC-3′); Gli1 (forward, 5′-TCCTACCAGAGTCCCAAGTT-3′; reverse, 5′-CCCTATGTGAGCCCTATTT-3′); Gli2 (forward, 5′-ATGAAGCCTAACTCTTGAGGTCT-3′; reverse, 5′-AACCTGGAATCAGAATGTGCTC-3′); GAPDH (forward, 5′-CGACCACTTTGTCAAGCTCA-3′; reverse, 5′-CCCTGTTGCTGTAGCCAAAT-3′); SuFu (forward, 5′-TGGCACTACATCAGCTTCGG-3′; reverse, 5′-TCAACTCAAAGCCAAAACCAC-3′); and PTCH1 (forward, 5′-ACTTCAAGGGGTACGAGTATGT-3′; reverse, 5′-TGCGACACTCTGATGAACCAC-3′).
Statistical Analysis
Values are shown as the mean ± SD. Differences between the groups were evaluated using Student’s t-test or one-way analysis of variance (ANOVA). Spearman correlation was used to evaluate the relationship between SPOP expression and clinical pathologic features. The graphical presentations were prepared using the GraphPad Prism Software (La Jolla, USA). All the analyses were performed using IBM SPSS23.0 software (Armonk, NY, USA). Differences were considered significant if P < 0.05 (*P < 0.05, **P < 0.01, and ***P < 0.001).
Results
Expression And Clinical Relevance Of SPOP In Ovarian Cancer
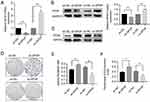
A total of 30 normal ovarian tissues and 55 epithelial ovarian cancer tissues were eligible for our study. We examined the expression level of SPOP in the normal ovarian and epithelial ovarian cancer tissues using immunohistochemical staining. SPOP was strongly expressed in most of the normal ovarian tissue samples but weakly in most of the cancer tissue samples (Figure 1A). Furthermore, although SPOP was localized in both the nucleus and cytoplasm of normal ovarian epithelial cells, it was mainly detected in the cytoplasm of epithelial ovarian cancer cells. We determined the SPOP staining score (from 0 to 12) of each section based on the intensity and area of SPOP protein expression. We found significant differences in the SPOP scores between normal ovarian tissues and epithelial ovarian cancer tissues. The SPOP score of normal ovarian epithelial tissues was much higher than that of epithelial ovarian cancer tissues (Figure 1B).
Although SPOP expression was low in epithelial ovarian cancer tissues, there were differences in SPOP levels among the samples. During the immunohistochemical SPOP scoring, we defined scores < 6 as low SPOP expression and those ≥ 6 as high expression. We further analyzed the correlation between the SPOP expression and clinicopathological features. As shown in Table 1, SPOP expression was inversely correlated with the FIGO stage and histopathological grade but not with the patient’s age or pathological type. This association between the SPOP expression and clinicopathological features indicates that SPOP is associated with epithelial ovarian cancer progression and malignancy. Lower SPOP levels were associated with advanced ovarian cancer and a higher degree of malignancy. Taken together, these results indicate that SPOP is a potential tumor suppressor in epithelial ovarian cancer.
 |
Table 1 Correlation Between SPOP Expression And Clinic Pathological Information In Epithelial Ovarian Cancer |
SPOP Inhibits Proliferation Of OVCAR-3 Ovarian Cancer Cells
To determine the role of SPOP in ovarian cancer, the human epithelial ovarian cancer cell line OVCAR-3 was infected with lentiviruses carrying an SPOP gene vector (ov-SPOP), SPOP knockdown vector (sh-SPOP), or a negative control vector (sh-NC or ov-NC). After screening for the infected cell lines, overexpression levels and knockdown efficiencies were determined by qRT-PCR and Western blotting (Figure 2A and B). In addition, the PCNA (proliferating cell nuclear antigen) level was found to decrease as the SPOP level increased, and the SPOP level decreased as the PCNA level increased (Figure 2C). In the colony formation assay, the clone formation rate of the ov-SPOP cell line was significantly lower than that of the ov-NC cell line. However, the clone formation rate of the sh-SPOP cell line was significantly higher than that of the sh-NC cell line (Figure 2D and E). Furthermore, the results of the EdU proliferation assay showed that the percentage of EdU-positive nuclei in the ov-SPOP cells was significantly lower than that in the ov-NC cells. The percentage of EdU-positive nuclei in the sh-SPOP cells was significantly higher than that in the sh-NC cells (Figures 2F and 3A). In conclusion, SPOP appears to have a suppressive effect on the proliferation and cloning ability of OVCAR-3 cells in vitro.
SPOP Induces Apoptosis In OVCAR-3 Cells
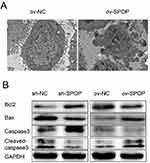
To determine whether SPOP had an effect on apoptosis in OVCAR-3 cells, we examined apoptotic bodies in the ov-SPOP and ov-NC cells using transmission electron microscopy (TEM). We observed that the ov-SPOP ovarian cancer cells contained apoptotic bodies unlike the ov-NC cells (Figure 4A). Next, we analyzed the levels of apoptosis-related proteins and found that overexpression of SPOP in OVCAR-3 cells upregulated the pro-apoptotic proteins cleaved-caspase3 and Bax while downregulating the apoptosis inhibitory protein Bcl2. In contrast, knocking down SPOP was found to downregulate cleaved-caspase3 and Bax while upregulating Bcl2 in OVCAR-3 cells (Figure 4B).
The Hh Signaling Pathway Is Involved In SPOP-Induced Apoptosis
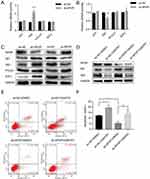
The qRT-PCR analysis revealed that the level of the Gli2 transcription factor of the Hh signaling pathway also changed according to the SPOP level. The down-regulation and up-regulation of SPOP significantly increased and decreased the Gli2 mRNA levels, respectively. However, Gli1, PTCH1, and SuFu mRNA levels did not change. These results suggested that SPOP might be involved in the Hh signaling pathway (Figure 5A and B). We further verified the SPOP-induced changes in the levels of the proteins related to the Hh signaling pathway. When we knocked down the SPOP, the levels of Gli1, Gli2, and SuFu proteins significantly increased, but there was no obvious change in the level of PTCH1. In contrast, as the SPOP protein level increased, the levels of Gli1, Gli2, and SuFu significantly decreased with no change on the PTCH1 level (Figure 5C). These results confirm that SPOP modulates the Hh signaling pathway. We then treated sh-SPOP and sh-NC cells with the inhibitor (GANT61) of the Hh signaling pathway to further determine whether SPOP induces apoptosis in ovarian cancer cells by inhibiting the Hh signaling pathway. The Western blotting results confirmed that GANT61 inhibited the expression of Gli1 and Gli2 (Figure 5D). Apoptosis was also quantified by flow cytometric analysis of the cells stained with annexin V-APC-A and DAPI. Treatment of sh-NC cells with GANT61 significantly increased the apoptotic rate relative to that observed with DMSO treatment. Treatment of sh-SPOP cells with GANT61 also somewhat induced apoptosis relative to that observed with DMSO treatment. Moreover, there was also a significant difference between the apoptotic rates of the sh-SPOP and sh-NC cells, showing that the inhibition of SPOP decreased the apoptosis rate than that of the negative control (Figure 5E and F). These results altogether indicate that SPOP protein promotes apoptosis in ovarian cancer cells by inhibiting the Hh signaling pathway.
Discussion
SPOP is a CUL3 ubiquitin ligase adaptor that promotes ubiquitination and degradation of its substrates, and it is involved in the regulation of a variety of signaling pathways. Studies on tumors have reported that SPOP may participate in the regulation of cell proliferation and apoptosis in a variety of tumors. Previously, we have found that the SPOP gene locus has a high-frequency deletion or loss of heterozygosity in ovarian cancer.22 However, the role of SPOP in ovarian cancer and its mechanism have remained unclear. In this study, we found differences in SPOP expression between 55 epithelial ovarian cancer cases and 30 normal ovarian samples. The significant decrease in SPOP expression in epithelial ovarian cancer tissues indicates that SPOP plays a role in the progression of ovarian cancer, and this conclusion is consistent with previous results. Correlation analysis revealed that SPOP expression was inversely correlated with the FIGO stage and histopathological grade. As the progression of ovarian tumors and the degree of malignancy increase, the expression of SPOP protein gradually decreases. This observation suggests that SPOP may be a suppressor of ovarian epithelial tumors. This effect was confirmed by in vitro experiments; we found that SPOP promoted apoptosis and inhibited proliferation in OVCAR-3 cells.
The Hh pathway exhibits elevated expression in ovarian cancer.34 Gli protein is the terminal effector in the Hh pathway, and as a transcription factor, it can regulate a variety of genes involved in cell proliferation, cell cycle, and apoptosis. SPOP has previously been shown to regulate the Hh signaling pathway by directly ubiquitinating Gli2 and Gli3 for proteasomal degradation.16,19,33 Moreover, SPOP can down-regulate SuFu in Drosophila by retaining the spliceosome factor Crooked neck (Crn).35 In this study, we found that SPOP regulated Gli2 at the transcriptional level since SPOP upregulation decreased the mRNA level of Gli2. Moreover, SPOP can downregulate Gli1 and SuFu proteins, the mechanism needs further research. The regulation of SPOP on Hh signaling pathway, its exact mechanism may depend on the cellular context, such as the type of cancer cells. It is conceivable that research on the regulation of the Hh signaling will provide new directions for cancer research and treatment.
Conclusion
Our study found that SPOP expression was significantly reduced in epithelial ovarian cancer tissues. In addition, increased SPOP expression suppressed cell proliferation and promoted apoptosis in the epithelial ovarian cancer cell line OVCAR-3. Moreover, SPOP promoted apoptosis by downregulating the Hh signaling pathway in OVCAR-3. All these data suggest that SPOP may play a key tumor suppressor role in epithelial ovarian cancer. This conclusion provides a new perspective to better understand the pathogenesis and therapeutic targets of ovarian cancer. However, further studies are required to clarify the mechanism of action by which SPOP regulates the Hh signaling pathway.
 |
Figure 3 Cell proliferation was analyzed by EdU proliferation assay. (A) EdU-positive nuclei indicate proliferating cells (scale bar, 1000 µm). |
Ethics statement
The use of OVCAR-3 cell line and human ovarian tissue sections are allowed by the Ethics Committee of the Second Affiliated Hospital of Chongqing Medical University. The committee approved our informed consent exemption because the tissue samples were left after the pathological examination of the previously operated patients and did not have any adverse effects on those patients.
Acknowledgments
We wish to acknowledge Professor Linghu Hua for the kind gift of the OVCAR-3 cell line. We are grateful to the College of Life Sciences of Chongqing Medical University for providing experimental equipment and technical support funded by the collective fund of the College of Life Sciences of Chongqing Medical University, China.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Rosen DG, Yang G, Liu G, et al. Ovarian cancer: pathology, biology, and disease models. Front Biosci (Landmark Ed). 2009;14:2089–2102. doi:10.2741/3364
3. Li C, Ao J, Fu J, et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30(42):4350–4364. doi:10.1038/onc.2011.151
4. Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–689. doi:10.1038/ng.2279
5. Le Gallo M, O’Hara AJ, Rudd ML, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44(12):1310–1315. doi:10.1038/ng.2455
6. Genschik P, Sumara I, Lechner E. The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. Embo J. 2013;32(17):2307–2320. doi:10.1038/emboj.2013.173
7. Zhuang M, Calabrese MF, Liu J, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36(1):39–50. doi:10.1016/j.molcel.2009.09.022
8. Wei X, Fried J, Li Y, et al. Functional roles of Speckle-Type Poz (SPOP) protein in genomic stability. J Cancer. 2018;9(18):3257–3262. doi:10.7150/jca.25930
9. An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6(4):657–669. doi:10.1016/j.celrep.2014.01.013
10. Geng C, He B, Xu L, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci U S A. 2013;110(17):6997–7002. doi:10.1073/pnas.1304502110
11. An J, Ren S, Murphy SJ, et al. Truncated ERG oncoproteins from TMPRSS2-ERG fusions are resistant to SPOP-Mediated proteasome degradation. Mol Cell. 2015;59(6):904–916. doi:10.1016/j.molcel.2015.07.025
12. Zhang P, Gao K, Jin X, et al. Endometrial cancer-associated mutants of SPOP are defective in regulating estrogen receptor-alpha protein turnover. Cell Death Dis. 2015;6:e1687. doi:10.1038/cddis.2015.47
13. Gao K, Jin X, Tang Y, et al. Tumor suppressor SPOP mediates the proteasomal degradation of progesterone receptors (PRs) in breast cancer cells. Am J Cancer Res. 2015;5(10):3210–3220.
14. Blattner M, Liu D, Robinson BD, et al. SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell. 2017;31(3):436–451. doi:10.1016/j.ccell.2017.02.004
15. Zhang L, Peng S, Dai X, et al. Tumor suppressor SPOP ubiquitinates and degrades EglN2 to compromise growth of prostate cancer cells. Cancer Lett. 2017;390:11–20. doi:10.1016/j.canlet.2017.01.003
16. Zeng C, Wang Y, Lu Q, et al. SPOP suppresses tumorigenesis by regulating Hedgehog/Gli2 signaling pathway in gastric cancer. J Exp Clin Cancer Res. 2014;33:75. doi:10.1186/s13046-014-0075-8
17. Ji P, Liang S, Li P, et al. Speckle-type POZ protein suppresses hepatocellular carcinoma cell migration and invasion via ubiquitin-dependent proteolysis of SUMO1/sentrin specific peptidase 7. Biochem Biophys Res Commun. 2018;502(1):30–42. doi:10.1016/j.bbrc.2018.05.115
18. Ding D, Song T, Jun W, Tan Z, Fang J. Decreased expression of the SPOP gene is associated with poor prognosis in glioma. Int J Oncol. 2015;46(1):333–341. doi:10.3892/ijo.2014.2729
19. Zhi X, Tao J, Zhang L, Tao R, Ma L, Qin J. Silencing speckle-type POZ protein by promoter hypermethylation decreases cell apoptosis through upregulating hedgehog signaling pathway in colorectal cancer. Cell Death Dis. 2016;7(12):e2569. doi:10.1038/cddis.2016.435
20. Chen L, Pei H, Lu SJ, et al. SPOP suppresses osteosarcoma invasion via PI3K/AKT/NF-kappaB signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22(3):609–615. doi:10.26355/eurrev_201802_14275
21. Li G, Ci W, Karmakar S, et al. SPOP promotes tumorigenesis by acting as a key regulatory hub in kidney cancer. Cancer Cell. 2014;25(4):455–468. doi:10.1016/j.ccr.2014.02.007
22. Hu X, Yang Z, Zeng M, et al. Speckle-type POZ (pox virus and zinc finger protein) protein gene deletion in ovarian cancer: fluorescence in situ hybridization analysis of a tissue microarray. Oncol Lett. 2016;12(1):658–662. doi:10.3892/ol.2016.4643
23. Briscoe J, Therond PP. The mechanisms of hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14(7):416–429. doi:10.1038/nrm3598
24. Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15(23):3059–3087. doi:10.1101/gad.938601
25. Reda J, Vachtenheim J, Vlckova K, Horak P, Vachtenheim J
26. Klieser E, Swierczynski S, Mayr C, et al. Differential role of hedgehog signaling in human pancreatic (patho-) physiology: an up to date review. World J Gastrointest Pathophysiol. 2016;7(2):199–210. doi:10.4291/wjgp.v7.i2.199
27. Liao X, Siu MK, Au CW, et al. Aberrant activation of hedgehog signaling pathway in ovarian cancers: effect on prognosis, cell invasion and differentiation. Carcinogenesis. 2009;30(1):131–140. doi:10.1093/carcin/bgn230
28. Chen Q, Gao G, Luo S. Hedgehog signaling pathway and ovarian cancer. Chin J Cancer Res. 2013;25(3):346–353. doi:10.3978/j.issn.1000-9604.2013.06.04
29. Li H, Li J, Feng L. Hedgehog signaling pathway as a therapeutic target for ovarian cancer. Cancer Epidemiol. 2016;40:152–157. doi:10.1016/j.canep.2015.11.014
30. Scales SJ, de Sauvage FJ. Mechanisms of hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci. 2009;30(6):303–312. doi:10.1016/j.tips.2009.03.007
31. Teglund S, Toftgard R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010;1805(2):181–208. doi:10.1016/j.bbcan.2010.01.003
32. Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10(6):719–729. doi:10.1016/j.devcel.2006.05.004
33. Zhang Q, Shi Q, Chen Y, et al. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2009;106(50):21191–21196. doi:10.1073/pnas.0912008106
34. Szkandera J, Kiesslich T, Haybaeck J, Gerger A, Pichler M. Hedgehog signaling pathway in ovarian cancer. Int J Mol Sci. 2013;14(1):1179–1196. doi:10.3390/ijms14011179
35. Liu C, Zhou Z, Yao X, et al. Hedgehog signaling downregulates suppressor of fused through the HIB/SPOP-Crn axis in drosophila. Cell Res. 2014;24(5):595–609. doi:10.1038/cr.2014.29
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.