Back to Journals » International Journal of Nanomedicine » Volume 12
Species-specific identification of collagen components in Colla corii asini using a nano-liquid chromatography tandem mass spectrometry proteomics approach
Authors Li X, Shi F, Gong L, Hang B, Li D, Chi L
Received 12 March 2017
Accepted for publication 23 May 2017
Published 15 June 2017 Volume 2017:12 Pages 4443—4454
DOI https://doi.org/10.2147/IJN.S136819
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Linlin Sun
Xue Li,1 Feng Shi,2 Liping Gong,2 Baojian Hang,2 Daoyuan Li,1 Lianli Chi1
1National Glycoengineering Research Center, Shandong Provincial Key Laboratory of Carbohydrate Chemistry and Glycobiology, and State Key Laboratory of Microbial Technology, Shandong University, Jinan, Shandong, People’s Republic of China; 2Scientific Research Division, Shandong Institute for Food and Drug Control, Jinan, Shandong, People’s Republic of China
Abstract: Colla corii asini (CCA) is a protein-based traditional Chinese medicine made from donkey skins. Because it has the ability to nourish blood, its demand is increasing rapidly. The shortage of donkey skins increases the risk of the adulteration of CCA products with other animal skins. To ensure the drug efficacy and safety of CCA products, a proteomics technique was applied to reveal proteins in the skins of donkey, horse, cattle, and pig. Species-specific peptides for each animal species were predicted using bioinformatics, and their presence in the skins and gelatin samples was examined by nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/MS). One unique marker peptide for each animal species was selected to develop an LC-MS/MS multiple reaction monitoring method. The capability of this method to identify donkey, horse, cattle, and pig materials was demonstrated by analyzing in-house-made donkey gelatins containing different amounts of other animal skins and commercial CCA products. The adulteration of non-donkey species could be sensitively detected at a low level of 0.5%. Hybrid animals, such as mules and hinnies, were also differentiated from donkeys. We provide a practical tool for the quality control of CCA products. The strategy can also be used to study other important traditional Chinese medicines which contain animal proteins.
Keywords: Colla corii asini, collagen, proteomics, adulteration, mass spectrometry
Introduction
Colla corii asini (CCA), also called Ejiao or donkey-hide gelatin, is a traditional Chinese medicine which is prepared from donkey (Equus asinus) skin.1 Collagen is believed to be the main active component in CCA, which is widely used to nourish the blood, optimize the immune response, improve the metabolic balance, delay senescence, and treat gynecologic diseases.2–6 Because CCA is very effective in practice, in 2015, its yearly production and market sales increased to approximately 6,000 tons and 1.5 billion US dollars, respectively. However, because donkeys are not major livestock animals, the increasing demand for CCA will inevitably cause a shortage of materials. Substitutions of donkey skins with those from other animals, such as cattle, horses, mules, and pigs, are often reported. Unlike donkey skin-made CCA, the efficacy and safety of which have been tested in human trials for over 2,000 years, the gelatin made from other animal species may cause severe problems.7 On the one hand, it could possess little to no effect compared to the authentic products. On the other hand, infectious risks are associated with some animal species.8 For example, cattle tissues cannot be used for the production of heparin, an anticoagulant drug, in many countries because of the potential for spongiform encephalopathy infection.9 Furthermore, other unknown side effects may occur if CCA is adulterated with non-donkey skins or tissues. To ensure the quality and safety of CCA products, a sensitive and reliable analytical approach is urgently needed to identify the animal species from which CCA is produced and detect the possible adulteration with other animal tissues.
Different analytical methods, including two-dimensional correlation infrared spectroscopy, near-infrared spectroscopy, and X-ray fluorescence, have been developed to detect counterfeit CCA.10 However, they are usually not sufficiently sensitive to detect partially adulterated skins from other animal species. Polymerase chain reaction (PCR) is a commonly used technique to track the source of an animal product or detect the possible contamination of other animal tissues. Whereas the PCR method’s effectiveness has been demonstrated in identifying CCA products,11 proteomics technique is emerging as an interesting tool to directly analyze the main active component in CCA, collagen proteins. Collagen accounts for over half of all extracellular proteins in the skin. It is an important structural protein and provides mechanical strength to connective tissues.12 It also functions in blood clotting, and mutated collagen can lead to various diseases.13 Collagen is a complex family of proteins with more than 20 types. Collagen type I is the main component in skin. It forms a triple helix consisting of three α polypeptide chains with more than 1,000 amino acid residues. Two chains called α 1(I) are the same, and the third is called α 2(I). The constitutional repeating unit in collagen is G-X-Y, where G is glycine and X and Y can be any amino acid residue except tryptophan. Most frequently, X is a proline residue, and Y is a hydroxyproline residue.14 The post-translational modification (PTM) of proline residues to hydroxyprolines by prolyl hydroxylase commonly occurs in collagen. The hydroxylation then contributes to the formation of hydrogen bonds and stabilizes the triple helix structure. Hydroxylation can also occur on lysine residues to form hydroxylysines.15 In addition to collagen type I, many other proteins are present in the skin, making the analysis difficult. Additionally, the protein homology among the herbivorous mammals makes protein-based species identification more challenging.
Proteomics is emerging as a powerful method to profile the total proteins in biological samples.16 Tens of thousands of proteins can be revealed simultaneously from cell culture or from animal/human tissue by liquid chromatography-tandem mass spectrometry (LC-MS/MS) and a subsequent database search.17,18 The subtle changes in a specific protein (eg, a single amino acid residue mutation or modification) can be determined ambiguously by high-resolution MS and MS/MS.19 Currently, LC-MS/MS proteomics methods have been widely used in the biopharmaceutical industry to ensure the appropriate structure, sequence, and PTMs of active ingredient proteins and for the detection of trace contaminant proteins from host cells.20 Some attempts to use proteomics to analyze CCA products have been previously reported.21,22 These methods are based on the clustering analysis of LC-MS/MS data. Another strategy for developing proteomic method to identify animal tissues is predicting the potential marker peptides by bioinformatics analysis, and validating them by LC-MS/MS experiments. When adulteration of other animal tissues is suspected, this target-orientated strategy usually works out more promptly. In this study, we demonstrated the usage of this new proteomics strategy to determine the original animal species used in CCA products. Bioinformatics was used to predict the potential marker peptides which are specific to each animal species. The presence of these marker peptides in raw skins of different animals, and in gelatin prepared from these skins was then tested by trypsin digestion and LC-MS/MS. The ability to sensitively detect adulteration in CCA using this method was also determined.
Materials and methods
Ethics
This study has been approved by the Ethics Committee of Qilu Hospital of Shandong University. All experiments were undertaken in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals with the approval of the Scientific Investigation Board of Shandong University School of Medicine, Jinan, Shandong Province, People’s Republic of China.
Chemicals and reagents
Trypsin (sequencing grade) was obtained from Promega (Fitchburg, WI, USA). Ultrafiltration units with a 10 kDa molecular weight cut-off (MWCO) were purchased from Sartorius (Goettingen, Germany). Guanidine hydrochloride, DL-dithiothreitol and iodoacetamide were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). Formic acid, acetonitrile (high-performance liquid chromatography [HPLC]-grade), and water (HPLC-grade) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). All other chemicals and reagents were of the highest grade available. Marker peptide standards were synthesized by Chinapeptides (Shanghai, People’s Republic of China). Skin and commercial CCA samples were collected by the Shandong Institute for Food and Drug Control. In-house gelatins were prepared by soaking skin pieces with 1% Na2CO3 solution at 60°C for 10 min and decocting at 120°C for 4 h in water.
LC-MS/MS shotgun analysis
Fresh skin samples were washed, dehaired, and cut into small pieces. Samples were then dehydrated by lyophilization and degreased with acetone. Samples were soaked in water, vortexed, and minced until the texture became loose. They were lyophilized once more and reconstituted to a concentration of 5 μg/μL. A 300 μL aliquot was mixed with 450 μL of denaturing buffer (0.5 M Tris-HCl, 2.75 mM EDTA, 6 M guanidine hydrochloride, pH 8.1). Proteins were reduced by incubation with 90 μL of 1 M DL-dithiothreitol at 37°C for 2 h and alkylated with 150 μL of 1 M iodoacetamide for 1 h in the dark. Reaction reagents were removed by ultrafiltration (MWCO 10 kDa). Samples were digested with trypsin at an enzyme-to-protein ratio of 1/50 (w/w) at 37°C for 24 h, and peptides were recovered by ultrafiltration (MWCO 10 kDa).
Dried gelatin samples were grinded into powder and suspended in water. After centrifugation at 12,000 rpm for 10 min, three layers formed. The middle layer was collected and degreased with n-hexane by centrifugation at 12,000 rpm for 10 min. The aqueous layer was transferred to a new tube. Protein reduction, alkylation, and digestion followed the same procedures as the skin samples.
Nano-LC-MS/MS analysis was performed on a Thermo Easy-nanoLC and an Orbitrap Fusion mass spectrometer equipped with a nanospray ion source. Peptides were separated using a ReproSil-Pur C18-AQ trap column (0.2 mm×3.5 cm) and a ReproSil-Pur C18-AQ analytical column (75 μm×25 cm). Mobile phase A was 0.1% formic acid in 2% acetonitrile, and mobile phase B was 0.1% formic acid in 98% acetonitrile. A step gradient of B 0–7% for 15 min, 7%–22% for 55 min, 22%–35% for 20 min, and 35%–90% for 5 min was used. The flow rate was set at 300 nL/min. The MS/MS parameters were set as follows: capillary voltage, 2.1 kV; ion transfer tube temperature, 275°C; ion spray voltage, 2,200 V; Orbitrap resolution, 120,000; scan range, 350 to 1,550; quadrupole isolation window, 2; and collision energy, 35%. The protein search was performed using the Thermo Scientific Proteome Discoverer 2.1 software.
Bioinformatics
Collagen sequences, including both COL1A1 and COL1A2 from donkey, horse, and cattle and COL1A2 from pig, were downloaded from the National Center for Biotechnology Information (NCBI) website (https://www.ncbi.nlm.nih.gov/protein/, GI: 221665286, 953866084, 75775290, 221665294, 149705490, 151555719, and 343887367), which constituted the database we built. The protein sequences from different animal species were compared using the BioEdit Sequence Alignment Editor software. The unique amino acid residues were spotted, and the corresponding tryptic peptides containing these amino acid residues were integrated into a small database. Next, the coverage of these peptides in the nano-LC-MS/MS experiments was examined by searching against only COL1A1 and COL1A2 from all four animal species. The MS/MS spectra of the matched peptides were examined using the Thermo Xcalibur software 2.2.
Database searching
The coverage and identified peptides were shown with Thermo Scientific Proteome Discoverer 2.1 software. The general settings were listed as follows: mass analyzer was ion trap mass spectrometer (ITMS), MS order was MS2, activation type was collision induced dissociation (CID), polarity mode was positive, the database was theoretical collagen sequencing (COL1A1 and COL1A2) of all species studied, enzyme name was trypsin, precursor mass tolerance was 10 ppm, fragment mass tolerance was 0.8 Da, dynamic modifications (peptide terminus) were oxidation/+15.995 Da(M), pro-hydroxypro/+15.995 Da(P), lys-hydroxy-lys/+15.995 Da(K), dynamic modifications (protein terminus) were acetyl/+42.011 Da(N-terminus), static modifications were carbamidomethyl/+57.021 Da(C).
LC-MS/MS multiple reaction monitoring (MRM) analysis
The reduction and alkylation steps were omitted. Skin or gelatin samples were digested with trypsin (1 μg/μL in 1% NH4HCO3) at 37°C for 24 h. The LC-MS/MS MRM analysis was performed using Waters ACQUITY Ultra-Performance LC system and a Quattro Premier XE mass spectrometer equipped with an electrospray ionization (ESI) source (Manchester, UK). The column was an ACQUITY UPLC BEH C18 (1.7 μm, 2.1×100 mm). Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile. The gradient was mobile phase B 4% for 3 min, 4%–8% for 5 min, 8%–50% for 2 min, and 50% for 2 min. The flow rate was set at 0.3 mL/min. The autosampler and column temperature was maintained at 10°C and 40°C, respectively. The mass spectrometer was in MRM mode, and the cone voltage was set at 20 V, the source and desolvation temperature were set at 150°C and 400°C, respectively. The collision energy for each marker peptide was optimized using the corresponding synthetic peptides (10 for DM1, 15 for HM1, 18 for CM2 and PM1). The MarkerLynx software version 4.1 was used to monitor every MRM transition spectrogram and the peak intensities.
Results and discussion
Study design
Shotgun proteomics is becoming a widely adapted technique for characterizing complicated protein mixtures. A typical shotgun analysis incorporates protein extraction, proteolytic digestion, LC-MS/MS analysis, and a database search to identify as many proteins as possible in a given sample. However, the proteins in skin are highly homologous among different mammal species. As a result, the shotgun approach may reveal the same set of proteins and cannot distinguish the identities of skin or gelatin samples. In this study, an integrated shotgun and targeted proteomics strategy was developed, as presented in Figure 1. Shotgun analysis was first used to profile the skin proteins of donkey, cattle, horse, and pig. The predominant proteins were recovered and their theoretical sequences were aligned using bioinformatics tools. All characteristic tryptic peptides were selected to establish a database. The shotgun LC-MS/MS experimental data were then re-examined by searching the peptide database against the parent proteins from all four animal species. The recovered peptides were considered as potential markers. Next, the intactness of these potential marker peptides from skin to gelatin was evaluated by performing the same experiment on the gelatin samples prepared from different animals. Four marker peptides representing donkey, cattle, horse, and pig were selected, and their suitability for identifying CCA products and detecting possible adulteration was evaluated by MRM.
Shotgun proteomics
Proteins were extracted from fresh skin pieces and digested with trypsin. Tryptic peptides were analyzed by nano-LC-MS/MS and the data were imported into Proteome Discoverer software to match the known proteins to corresponding species in the NCBI database. The number of identified proteins for each species was quite different, ranging from 918 to 2,184 (P<0.05). The volume of known proteins of each animal species in the database determined how many peptides could be interpreted in the shotgun experiment. However, the sequence coverage of the matched proteins was relatively low; most were lower than 50%, even for the high peptide-spectrum match (PSM) proteins. The hydroxylation of proline and lysine residues, either endogenous or artificially modified during the process, represents one possible reason for the low sequence coverage. After adding these two possible PTMs during the database search, the number of identified proteins (P<0.05) increased to 1,772 for donkey, 3,120 for horse, 2,787 for cattle, and 4,017 for pig. These results indicate that the proteins in mammal skins are heavily modified by hydroxylation. The variable peptides made the species identification more difficult. The sequence coverage of high PSM proteins also increased significantly to 60% or higher, making the majority of these protein sequences detectable. The identities of the top PSM proteins among the four animal species were essentially the same and were mainly collagens. Non-collagenous proteins, such as keratins, albumin, and fibrillin, were also detected. Collagens type I and type III were mainly distributed in the skin; however, the sequences of collagen type III of the four animal species examined here were not complete in the NCBI database. Additionally, it has been reported that the content of collagen type III in the skin decreases with age. Therefore, collagen type I was chosen to identify potential marker peptides.
Bioinformatics
Collagen type I consists of two subunits: COL1A1 and COL1A2. The NCBI database contains both COL1A1 and COL1A2 sequences from donkey, horse, and cattle but only the COL1A2 sequence from pig. The available COL1A1 and COL1A2 sequences from different animal species were aligned and compared using the BioEdit Sequence Alignment Editor software. As an example, the partial sequence alignments of COL1A1 of donkey, horse, and cattle as well as COL1A2 of donkey, horse, cattle, and pig are displayed in Figure 2. The identical amino acid residues among different species are represented in the same color, and discrepant amino acid residues are represented in different colors for easy identification. Ten amino acid residues located within this sequence were not identical among the different species, which yielded eleven potential marker tryptic peptides: 497GPTGEPGKPGDK508 for donkey; 422GASGPAGVR430 and 497GPSGEPGKPGDK508 for horse; 352GEGGPQGPR360, 497GPSGDPGK504, and 518GAPGPDGNNGAQGPPGLQGVQGGK541 for cattle; 412AGVMGPPGSR421, 422GPTGPAGVR430, 449GFPGSPGNVGPAGK462, 463EGPAGLPGIDGR474, and 497GPTGDPGK504 for pig. Using bioinformatics, a total of 31 potential marker peptides were obtained and are summarized in Table 1. Most potential marker peptides were from COL1A2. Because the database lacks information on pig COL1A1, the suitability of some peptides for differentiating the four animal species should be verified in future experiments.
Selection of marker peptides for each animal species
The presence of all theoretical marker peptides in skin samples was verified by LC-MS/MS experiments. The marker peptides for skin were selected based on two criteria. First, the peptide can be detected consistently during multiple repetitive runs. Second, the ideal length of the peptide should range from 7 to 15 amino acid residues, making it feasible for the MRM method. The selection of donkey skin marker peptides was performed as an example. Among the three theoretical marker peptides discovered by an informatics comparison, the peptide 497GPTGEPGKPGDK508 of COL1A2 (DM1 in Figure 3A) and 882VGPPGPSGNAGTPDPPGPVGK902 of COL1A1 were detected consistently from multiple skin samples. The peptide 179GHK181 of COL1A2 was too short to be recovered. DM1 was detected only in donkey skin samples, making it a good marker peptide for donkey skin. In the case of horse and pig skins, the unique peptides 422GASGPAGVR430 (HM1 in Figure 3B) and 422GPTGPAGVR430 (PM1 in Figure 3D) were detected, but in the other two animals, the corresponding peptide 422GATGPAGVR430 with a single amino acid residue difference (Figure 3E) was found. The peptide 352GEGGPQGPR360 (CM2 in Figure 3C) was the unique peptide for cattle skin. Six skin marker peptides (one for donkey, two for horse, two for cattle, and one for pig) were selected and are summarized in Table 2.
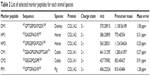  | Table 2 List of selected marker peptides for each animal species |
In the CCA manufacturing process, skin materials undergo very harsh conditions, including decoction with oil, sugar, liquor, and other excipients at a high temperature for a prolonged period of time. Therefore, proteins in the skin may be degraded. To ensure that the marker peptide sequences remained intact in the final products, gelatins made from donkey, horse, cattle, and pig skins were analyzed using trypsin digestion and LC-MS/MS analysis. The ions with m/z values corresponding to marker peptides were extracted from each LC-MS/MS run, and their m/z values, charge states, and retention times are summarized in Table 2. All marker peptides were detectable in their corresponding gelatins, but did not exist in CCAs produced from any other animal species. The hydroxylation modification of these peptides was also evaluated. By extracting the selected ions from the gelatin LC-MS/MS analysis, the peptide DM1 showed the co-existence of non-hydroxylated, singly hydroxylated, and doubly hydroxylated forms. The non-hydroxylated form accounts for over 99% of the total peak areas of three forms. No hydroxylated modification was detected for peptide HM1, CM2, and PM1. Therefore, one exclusive unmodified peptide for each species (DM1 for donkey, HM1 for horse, CM2 for cattle, and PM1 for pig) was used to develop the quantitation method.
In the Chinese Pharmacopoeia, the peptide GPPGAAGPPGPR is considered a marker peptide for authentic CCA products made from donkey skin.1 However, we found this peptide not only in donkey skin but also in horse skin (Figure 3F). Therefore, it should not be used to identify cross-species contamination, and a revision of this method in the Pharmacopoeia is recommended.
Development of the MRM quantitation method for marker peptides
MRM experiments were performed on a triple-quadrupole mass spectrometer. Typically, multiple transitions are required to quantify a peptide from complex samples, whereas a single transition may be sufficient for monitoring peptides from particular proteins with high abundance.23 Collagen is the dominant protein in animal skins and CCA products. Three or more transitions were tested for each marker peptide and the most abundant ones were selected in order to simplify the method. For example, transitions 427.76>457.25, 427.76>554.31, and 427.76>668.26 were tested for monitoring CM2, and 427.76>668.26 exhibited the most intensive signal. Four most intensive transitions representing four marker peptides were included in the LC-MS/MS MRM scans (380.70 [triply charged]>374.82 for DM1, 386.25>499.25 for HM1, 427.76>668.26 for CM2, and 406.55>556.81 for PM1). The MRM chromatograms of gelatins prepared from the four animal species are shown in Figure 4. The marker peptides DM1, HM1, CM2, and PM1 were eluted at 1.31 min, 2.55 min, 1.38 min, and 5.15 min, respectively. The identity of each CCA product was revealed unambiguously by the presence of corresponding marker peptides and the absence of marker peptides of other species.
Next, these four marker peptides were synthesized to develop a quantitation method. Solutions of these peptides with various concentrations, ranging from 0.1 μg/mL to 10 μg/mL, were prepared. All concentrations were analyzed in triplicate, and 1 μL of sample was injected for each LC-MS/MS MRM run. As shown in Figure 5, standard curves were plotted using the MRM peak area against the amount of sample injected. Good linearity was obtained, and all R2 values were greater than 0.99. The limit of detection – limit of quantitation of DM1, HM1, CM2, and PM1 was 1.04 pg – 3.47 pg, 6.17 pg – 20.56 pg, 4.05 pg – 13.50 pg, and 8.93 pg – 29.76 pg, respectively.
Detection of adulterated CCA products
To demonstrate the ability of this MRM method to identify adulterated CCA products with animal species other than donkey, the authentic donkey CCA gelatin was mixed with 0.1%, 0.5%, and 1% of horse, cattle, and pig gelatins, respectively. Sample preparation was simplified to adapt this method for use in routine applications. Triplicates were analyzed for each sample, and the representative MRM chromatograms are shown in Figure 6A–D. When no other animal gelatin was added, only the marker peptide for donkey was observed. The addition of gelatins from three other animal skins was easily detected at an amount over 0.5% of the total weight in repetitive runs. These results suggest that this method is highly sensitive and is able to detect a trace amount of adulterated animal material in CCA products.
Ten batches of commercial CCA products from different manufacturers were inspected using this new method. Eight of them were proven to be authentic products, as only the donkey marker peptide DM1 was detected in MRM analysis (Figure 6E). The presence of horse marker peptide HM1 was found in addition to DM1 in two batches of CCA products from the same manufacturer, which were suspected to be adulterated with horse skin materials (Figure 6F).
Identification of CCA adulterated with hybrid animal skins
Mules and hinnies are hybrid offspring of donkeys and horses. Their skins are also possibly adulterated materials in CCA because they are common domestic animals, are much larger in size than donkeys, and are more similar to donkeys than other animals. A mule is bred from a male donkey and a female horse, and a hinny is bred from a male horse and a female donkey. The identification of a hinny’s skin is extremely difficult because the mitochondrial DNA of a hinny is the same as that of a donkey, and cannot be differentiated by PCR. The gelatins made from mule skin and hinny skin were separately analyzed using the LC-MS/MS approach. Interestingly, both marker peptides (DM1 for donkey and HM1 for horse) were detected in these two samples. The hybrid animals, either mule or hinny, contained two sets of proteins from both parents, and their presence was disclosed by the LC-MS/MS method when they were added to authentic CCA products.
Conclusion
Traditional Chinese medicine is an invaluable resource for modern health care and the pharmaceutical industry. The development of an anti-malarial drug, artemisinin, is a famous example. Tremendous effort is being devoted to studies on traditional Chinese medicines with modern technologies and methodologies; however, most focus on small plant-derived compounds. As recently as a few years ago, protein-based large biomolecule drugs have taken the lead over small molecules in the global pharmaceutical market. However, the structures and mechanisms of protein components in traditional Chinese medicines are extremely difficult to study, and quality control is also very challenging. CCA is an example of a traditional Chinese medicine consisting of complicated protein components. Herein, we established a highly sensitive and specific LC-MS/MS MRM approach to trace animal species in skin materials. The adulteration of non-authentic skin materials can be easily detected at a very low level. This novel method can be adapted for the routine analysis of skin or gelatin samples, as the sample processing steps were simplified and a relatively inexpensive mass spectrometer is required. The strategy of combining bioinformatics and shotgun proteomics can be applied to a wide spectrum of important traditional Chinese medicines which contain multiple protein components.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (21472115).
Disclosure
The authors report no conflicts of interest in this work.
References
Chinese Pharmacopoeia Committee. Chinese Pharmacopoeia. Vol I. 2015 ed. Beijing: Chinese Medical Science and Technology Press; 2015. | ||
Kumeta Y, Maruyama T, Asama H, Yamamoto Y, Hakamatsuka T, Goda Y. Species identification of Asini Corii Collas (donkey glue) by PCR amplification of cytochrome b gene. J Nat Med. 2014;68(1):181–185. | ||
Li Y, He H, Yang L, Li X, Li D, Luo S. Therapeutic effect of Colla corii asini on improving anemia and hemoglobin compositions in pregnant women with thalassemia. Int J Hematol. 2016;104(5):559–565. | ||
Wu H, Ren C, Yang F, Qin Y, Zhang Y, Liu J. Extraction and identification of collagen-derived peptides with hematopoietic activity from Colla Corii Asini. J Ethnopharmacol. 2016;182:129–136. | ||
Shen L, Chen H, Zhu Q, et al. Identification of bioactive ingredients with immuno-enhancement and anti-oxidative effects from Fufang-Ejiao-Syrup by LC-MS(n) combined with bioassays. J Pharm Biomed Anal. 2016;117:363–371. | ||
Wang D, Liu M, Cao J, et al. Effect of Colla corii asini (E’jiao) on D-galactose induced aging mice. Biol Pharm Bull. 2012;35(12):2128–2132. | ||
Gould J, Callis CM, Dolan DG, Stanard B, Weideman PA. Special endpoint and product specific considerations in pharmaceutical acceptable daily exposure derivation. Regul Toxicol Pharmacol. 2016;79 (Suppl 1):S79–S93. | ||
Lv P, Zhao Y, Qi F, et al. Authentication of equine DNA from highly processed donkey-hide glue (Colla Corii Asini) using SINE element. J Food Drug Anal. 2011;19(2):123–130. | ||
Guan Y, Xu X, Liu X, et al. Comparison of low-molecular-weight heparins prepared from bovine lung heparin and porcine intestine heparin. J Pharm Sci. 2016;105(6):1843–1850. | ||
Liu Y, Zhang G, Sun S, Noda I. Study on similar traditional Chinese medicines cornu Cervi pantotrichum, cornu Cervi and cornu Cervi degelatinatum by FT-IR and 2D-IR correlation spectroscopy. J Pharm Biomed Anal. 2010;52(4):631–635. | ||
Fumière O, Dubois M, Baeten V, von Holst C, Berben G. Effective PCR detection of animal species in highly processed animal byproducts and compound feeds. Anal Bioanal Chem. 2006;385(6):1045–1054. | ||
Gelse K, Pöschl E, Aigner T. Collagens – structure, function, and biosynthesis. Adv Drug Deliv Rev. 2003;55(12):1531–1546. | ||
Lohmann K, Schlicht F, Svetel M, et al. The role of mutations in COL6A3 in isolated dystonia. J Neurol. 2016;263(4):730–734. | ||
Pawelec KM, Best SM, Cameron RE. Collagen: a network for regenerative medicine. J Mater Chem B Mater Biol Med. 2016;4(40):6484–6496. | ||
Rodriguez-Pascual F, Slatter DA. Collagen cross-linking: insights on the evolution of metazoan extracellular matrix. Sci Rep. 2016;6:37374. | ||
Kayili HM, Salih B. Fast and efficient proteolysis by reusable pepsin-encapsulated magnetic sol-gel material for mass spectrometry-based proteomics applications. Talanta. 2016;155:78–86. | ||
Pease BN, Huttlin EL, Jedrychowski MP, et al. Global analysis of protein expression and phosphorylation of three stages of Plasmodium falciparum intraerythrocytic development. J Proteome Res. 2013;12(9):4028–4045. | ||
Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513(7518):382–387. | ||
Zielinska DF, Gnad F, Wiśniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141(5):897–907. | ||
Doneanu CE, Xenopoulos A, Fadgen K, et al. Analysis of host-cell proteins in biotherapeutic proteins by comprehensive online two-dimensional liquid chromatography/mass spectrometry. MAbs. 2012;4(1):24–44. | ||
Cheng XL, Wei F, Xiao XY, et al. Identification of five gelatins by ultra performance liquid chromatography/time-of-flight mass spectrometry (UPLC/Q-TOF-MS) using principal component analysis. J Pharm Biomed Anal. 2012;62:191–195. | ||
Yang H, Shen Y, Xu Y, et al. A novel strategy for the discrimination of gelatinous Chinese medicines based on enzymatic digestion followed by nano-flow liquid chromatography in tandem with orbitrap mass spectrum detection. Int J Nanomedicine. 2015;10:4947–4955. | ||
Mead JA, Bianco L, Ottone V, et al. MRMaid, the web-based tool for designing multiple reaction monitoring (MRM) transitions. Mol Cell Proteomics. 2009;8(4):696–705. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







