Back to Journals » Cancer Management and Research » Volume 12
SP1-Induced Upregulation of lncRNA LINC00514 Promotes Tumor Proliferation and Metastasis in Osteosarcoma by Regulating miR-708
Authors Mi LD, Sun CX, He SW, Du GY
Received 15 December 2019
Accepted for publication 31 March 2020
Published 11 May 2020 Volume 2020:12 Pages 3311—3322
DOI https://doi.org/10.2147/CMAR.S242464
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Li-Dong Mi, Chuan-Xiu Sun, Sheng-Wei He, Guang-Yu Du
Orthopeadic Surgery, Zuanshiwan Hospital District of the Second Hospital of Dalian Medical University, Dalian, Liaoning 116031, People’s Republic of China
Correspondence: Chuan-Xiu Sun
Orthopeadic Surgery, Zuanshiwan Hospital District of the Second Hospital of Dalian Medical University, Dalian, Liaoning 116031, People’s Republic of China
Email [email protected]
Background: Growing studies have suggested the dysregulation of long non-coding RNAs (lncRNAs) in several tumors, including osteosarcoma (OS). However, limited studies report metastasis-associated lncRNAs in OS. Our present study aimed to explore the roles of lncRNA LINC00514 (LINC00514) in OS.
Materials and Methods: The LINC00514 expression was measured using qPCR assays in OS tissues and cell lines. The clinical significance of LINC00514 expression in OS patients was analyzed using chi-square test, Kaplan–Meier assays and multivariate analysis. The possible effects of LINC00514 in tumor cellular progression were determined using a series of functional assays. The mechanisms of LINC00514 action were explored through bioinformatics, luciferase reporter assays and RT-PCR assays. The mechanisms involved the upregulation of LINC00514 expression in OS were determined using luciferase reporter and chromatin immunoprecipitation (ChIP) assays.
Results: We showed that LINC00514 expressions were distinctly upregulated in both OS tissues and cell lines, especially in advanced cases. High levels of LINC0051 were positively correlated with advanced tumor stages, distant metastasis, and reduced survival of patients with OS. Functional experiments indicated that silencing of LINC00514 suppressed the ability of cell growth, colony formation and metastasis, whereas promoted cell apoptosis in vitro. Mechanistic investigation revealed that LINC00514 could directly bind to miR-708 and effectively serve as a ceRNA for miR-708. In addition, LINC00514 was upregulated by the transcription factor SP1.
Conclusion: Our findings revealed SP1-induced upregulation of LINC00514 as an oncogene in OS through competitively binding to miR-708, suggesting that there are potential diagnostic and treatment values of LINC00514 in OS.
Keywords: LncRNA LINC00514, miR-708, osteosarcoma, prognosis, SP1, metastasis
Introduction
Osteosarcoma (OS) is one of the most common primary malignant tumors in children and young adults, and this tumor usually originates in the metaphysis of the long bones.1,2 The approximate incidence rate worldwide is 4.5/million/year, with a peak incidence at the age of 14–20.3 Although treatments and perioperative management have evolved in the past ten years with recent advancements in diagnosis progress, surgical procedures and the use of adjuvant chemotherapy, OS remains an extremely high morbidity and mortality.4,5 Although more and more tumor-related regulators have been identified, almost no commonly-accepted markers have been established for the clinical application.6–8 Thus, it is urgent to make clear the key molecule involved in the growth and metastasis of OS for the improvement of early detection and targeted treatment of OS.
With the advancement of next-generation sequencing methods, more and more dysregulated noncoding RNAs are identified in human tissues and they are confirmed to be frequently transcribed in the genome.9 Long noncoding RNAs (lncRNAs) are RNAs with more than 200 nucleotides in length and lack abilities of protein coding due to no functional open reading frame.10 In recent years, emerging evidences reveal that lncRNAs act as novel modulators of gene expressions via a series of mechanisms involved in epigenetic modification.11,12 Of note, increasing lncRNAs are demonstrated to be abnormally expressed in various tumor tissues, and some of lncRNAs functioning as oncogenes or tumor suppressors in particular conditions have been functionally characterized.13–15 In addition, the critical roles of lncRNAs in tumor progression highlighted the clinical application of lncRNAs used as novel cancer biomarkers.16,17
Recently, more and more abnormally expressed lncRNAs were demonstrated by bioinformatics analysis and various cell experiments using RT-PCR. However, the molecular mechanism involved in the abnormal expressions of lncRNAs remained largely unclear. Emerged evidences indicated that transcription factors have a central role in the transcription of genes, which highlighted the potential function of transcription factors as novel modulators in the expression of lncRNAs.18,19 In addition, several transcription factors such as SP1, STAT1 and STAT3 have been demonstrated to display functional roles in the regulation of lncRNAs.20–22
In this study, we identified a new c-related lncRNA, LINC00514 which was firstly functionally identified in papillary thyroid cancer.23 We firstly provided evidence that LINC00514 levels were upregulated in OS and predicted a poor clinical outcome. Further experiments indicated that overexpression of LINC00514 was induced by SP1. Moreover, we performed functional assays and mechanism experiments to explore the potential functions of LINC00514 in OS cells and the relative mechanisms. Overall, our findings provided a novel clue for the discovery of cancer biomarkers and therapeutic targets for OS patients.
Materials and Methods
Clinical Samples
OS specimens and adjacent normal tissues were acquired from 107 patients with OS that were enrolled in the Zuanshiwan Hospital District of The Second Hospital of Dalian Medical University from March 2010 to September 2013. The samples were collected with the written informed consents of the patients, and this study was approved by the ethics committee of Zuanshiwan Hospital District of The Second Hospital of Dalian Medical University. Each specimen after collecting were frozen in liquid nitrogen.
Cell Transfection
We bought five OS tumor cell lines (143B, Saos-2, MG63, U2OS, and HOS cells) and hFOB1.19 cells (as control cells) from Feijun Biological company (Changsha, Hunan, China). The cells were routinely cultured in RPMI-1640 media containing 10% FBS in a 5% CO2 incubator with 37 °C. The siRNAs targeting LINC00514 (lnc-siRNA-1, lnc-siRNA-2) and SP1 (si-SP1), control siRNAs (NC-siRNA, si-NC), miRNA mimics and inhibitors were all bought from Shenggong Biological company (Songjiang, Shanghai, China). The pcDNA3.1-SP1 and LINC00514 overexpressing plasmids (ov-LINC00514) were constructed by Laidun Biological company (Qingdao, Shandong, China). Lipofectamine 2000 reagent kits (Kefeng, Xiamen, Fujian, China) were used for cell transfection in accordance with the kits’ protocols.
Real-Time PCR
Trizol reagents (BoYuan, Ningbo, Zhejiang, China) were employed for extracting the total RNAs form OS samples or cells. ND-1000 UV (Thermo Scientific, Waltham, MA, USA) apparatus was used to identify the concentrations of total RNAs. The extracted RNAs were then subjected to reverse transcription using cDNA synthesis kits (TAKARA, Dalian, Liaoning, China). Then, qPCR analyses for LINC00514 and SP1 were carried out using SYBR Green qPCR kits (Kaijie, Suzhou, Jiangsu, China), and the reaction was operated on the basis of the kits’ protocols. For miR-708 detection, Transgen two-step miRNA qPCR kits (Longjun, Chengdu, Sichuan, China) were employed in accordance with the kits’ protocols. GAPDH was used as an internal control of LINC00514 and SP1 detection, while U6 was used as the internal control of miR-708 detection. Fold change was calculated using 2−ΔΔCt method. The primers are described in Table 1.
 |
Table 1 Sequences of Primers Used in This Study |
Western Blot
Total proteins of OS cells after treatment were lysed by RIPA buffer (Hongjun, Changsha, Hunan). The protein concentrations were then determined using the Pierce BCA kits (Dongkun, Chengdu, Sichuan, China). Thereafter, proteins were separated with 10% SDS-PAGE, followed being transferred onto PVDF membranes. After being blocked using 5% BSA, primary antibodies were used for probing the membranes for 12 h at 4 °C. After washing using TBST buffer, corresponding secondary antibodies were used for incubation with the membranes. The protein blots were visualized by applying ECL kits (Tengrui, Changsha, Hunan, China). GAPDH was used as the loading control. The primary antibodies against caspase 3 and caspase 9 were bought from PTG technology (Wuhan, Hubei, China).
Cell Viability Detection
The cellular proliferation was determined using CCK-8 kits (Weiteng, Shenzhen, Guangdong, China). After transfection with LINC00514 siRNAs, OS cells were placed into plates (96‐well; 2 × 103 cells per well). The cells were kept in a culture condition (5% CO2, 37°C) for the indicated time. At 24, 48, 72, or 96 h (1–4 days) after seeding, CCK‐8 reagents (10 μL) were put into each well, followed by 2.5 h of incubation at 37°C. Finally, the absorbance at 450 nm was evaluated using a microplate reader.
Colony Formation Assay
The transfected 143B and MG63 cells were respectively placed into plates (6-well; 800 cells per well), followed by being cultured for 2–3 weeks in complete media. After the colonies were visible, crystal violet (0.2%) was applied for treating these colonies for 15 min. After washing by PBS buffer, the colonies were photographed by a microscope.
TUNEL Assay
Promega TUNEL assay kits (Shengda, Hangzhou, Zhejiang, China) were used for cell apoptosis detection according to the kits’ protocols. In brief, the OS cells after treatment with corresponding siRNAs were placed into 48-well plates. After the cells attached the plates (more than 12 h), the cells were washed and treated with paraformaldehyde (4%). Subsequently, the cells were treated with proteinase K reagents and TdT buffer for 1 h at 37°C. After treating with TUNEL detection buffer, the cells were washed twice and the fluorescence was imaged by a fluorescence microscope.
Wound-Healing Assay
The cell migratory abilities were evaluated by wound-healing assays. In short, OS cells were treated with corresponding siRNAs, and 8–10 hours later, the cells were collected and re-plated in 12-well plates with high density. On the second day, the cell monolayers (near 100% cell confluence) were formed, and 200 μL tips were used for scraping the cells. The wound closures were imaged using a microscope at 0 h and 48 h after the cells were scraped.
Transwell Assay
To assess the invasive capacities of OS cells, 1.5 × 105 cells after treatment were added into the Corning transwell chambers (Xunfeng, Hefei, Anhui, China) coated with Matrigel. Twenty-four hours later, cells invasive to the bottom sides of the membranes were treated using formaldehyde (4%) and crystal violet (0.3%) for 15 min. After washing by PBS buffer, the colonies were photographed by a microscope.
Subcellular Fractionation Location Assay
The nuclei and cytoplasm of 143B cells were separated by employing Thermo Scientific Nuclei-Cytoplasm extracting kits (Hening, Ningbo, Zhejiang, China) in accordance with the kits’ protocols. The RNAs from nuclei or cytoplasm were isolated and subjected to qPCR detection as described above. U6 and GAPDH acted as markers of the nuclei and cytoplasm, respectively.
ChIP Assay
ChIP assays were carried out to evaluate the binding of SP1 and LINC00514 promoter. In brief, the cells after treatment were cross‐linked with formaldehyde (1%) and subsequently quenched by glycine (0.125 M), followed by being collected in ChIP lysis buffer after the cells were washed twice. Afterwards, the sonicators were used for shearing the DNAs into 200–400 bp. The chromatins were immune-precipitated by anti‐SP1 antibodies (4°C, 4 h) with IgG as a negative control, followed by adding Invitrogen protein G Sepharose (JunLong, Xiamen, Fujian, China). The mixture was incubated for 2 h at 4°C. The precipitated complex was rinsed twice, followed by adding elution buffer. Finally, qPCR was employed to analyze detect the immune-precipitated chromatin DNA.
Luciferase Reporter Assay
The binding site between LINC00514 and miR-708 was predicted using “miRDB” algorithm. The regions containing the predicted binding site was constructed into pGL3 luciferase reporter vector (LINC00514 wild-type). In addition, the wild-type of corresponding region was also constructed (LINC00514 mutant-type). The binding sites between LINC00514 and SP1 were predicted by “Jaspar” algorithm. Correspondingly, the sequence containing the predicted binding site 2 (PS2) was also constructed into pGL3 vector and the plasmid was named as PS2 WT (wild-type). In addition, PS2 mutant-type (PS2 MUT) reporters were also constructed. After transfection with corresponding reporter plasmids, the luciferase activities were evaluated by using Promega dual-luciferase reporter assay kits in accordance with the kits’ protocols (Shengda, Hangzhou, Zhejiang, China).
Statistical Analyses
SPSS 20.0 (SPSS, Chicago, IL, USA) software was used for statistical analyses. Student’s t test was used to examine pairwise comparisons and one-way ANOVA analysis was used to examine comparisons (more than two groups). Overall survival rates were analyzed using Kaplan–Meier methods and Log rank tests. Univariate and multivariate models were used examine the influence of related factors on patient survival. Differences were considered significant at p < 0.05.
Results
Aberrant Upregulation of LINC00514 Was Observed in OS Tissues and Cells
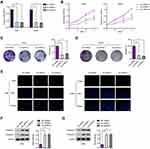
To determine whether LINC00514 was dysregulated in OS, we firstly examined LINC00514 expression in OS tissues and cells using qRT-PCR. Our results indicated that the expressions of LINC00514 were distinctly upregulated in OS specimens compared to matched normal specimens (Figure 1A, p < 0.01). In addition, patients with advanced stages displayed higher levels compared to other patients (Figure 1B), suggesting that higher levels of LINC00514 contributed to tumor progression. Then, we performed RT-PCR to detect the expression of LINC00514 in OS cells, finding that LINC00514 expression was distinctly higher in five OS cell lines than in hFOB1.19 (p < 0.01, Figure 1C). These results revealed that LINC00514 might play potential roles in the progression of OS.
Increased Expressions of LINC00514 Was Associated with the Poor Prognosis in OS
OS tissue samples were classified into the low-expressing group (n = 55) and the high-expressing group (n = 52) according to the median expression level of all OS samples. Table 2 showed the associations between several clinicopathological factors and LINC00514 levels. Our data indicated that high LINC00514 levels were positively correlated with tumor stage (p = 0.017) and distant metastasis (p = 0.031), suggesting that LINC00514 may contribute to clinical progression of this tumor. Thus, we wondered the possible correlation between LINC00514 expression and long-term overall. As shown in Figure 1D, we found that overall survival was higher in patients with high LINC00514 expression than in those with low LINC00514 expression (p = 0.0062). To further determine the prognostic values of LINC00514 in OS patients, univariate and multivariate assays were performed and the results revealed that LINC00514 (HR=2.896, 95% CI: 1.217–4.285, p =0.022) was an independent protective predictor of overall survival of OS patients (Table 3). Overall, our findings suggested LINC00514 as a novel biomarker for this tumor. However, more OS samples were needed to be analyzed for further confirmation of our results.
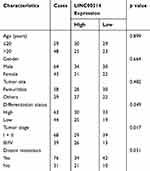 |
Table 2 Correlation Between LINC00514 Expression and Clinicopathological Characteristics in Osteosarcoma (n = 107) |
 |
Table 3 Univariate and Multivariate Analyses for Overall Survival in Osteosarcoma Patients |
LINC00514 Knockdown Suppressed OS Development in vitro
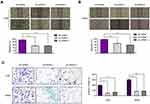
To explore the functional roles of LINC00514 in OS cells, we transfected siRNAs targeting LINC00514 into 143B and MG63 cells. Real-time PCR was conducted at 48 h post-transfection and indicated that LINC00514 siRNAs had a high efficiency of interference (Figure 2A). Thereafter, we sought to assess the effects of LINC00514 knockdown on cell proliferation. CCK-8 assays revealed that cellular growth was remarkably impaired in LINC00514 siRNAs-transfected OS cells (Figure 2B). Similarly, the data from colony formation assays demonstrated that LINC00514 deficiency reduced clonogenic survivals of OS cells (Figure 2C and D). Subsequently, to determine whether the effects of LINC00514 depletion on OS cell proliferation influenced apoptosis, TUNEL assays were performed. The results showed that the proportion of apoptotic cells following treatment with LINC00514 siRNAs was markedly increased compared with the controls (Figure 2E). Mechanically, data from Western blot validated that repressing the levels of LINC00514 remarkably accelerated the expression of caspase 3/9 in OS cells (Figure 2F and G). Overall, the data suggested that LINC00514 depletion was capable to depress OS development.
LINC00514 Inhibited the Metastatic Potentials of OS Cells
In spite of proliferation, metastasis is also an important feature of cancer cells. Therefore, we next attempted to investigate the influence of LINC00514 suppression on OS cell migration and invasion. First, we conducted wound-healing assays to evaluate the effects of LINC00514 downregulation on cell migration. As the data presented in Figure 3A and B, depression of LINC00514 notably elevated the velocity of cell movements. Afterwards, the transwell invasion assays demonstrated that cell invasion of OS cells was also suppressed by repressing LINC00514 expression (Figure 3C). Therefore, these data proved that depression of LINC00514 inhibited metastasis of OS cells.
LINC00514 Directly Targeted miR-708 and Acted as a miR-708 Sponge in OS Cells
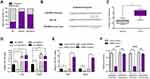
The previous data proved that LINC00514 deficiency depressed the malignant behaviors of OS cells. Since mount of studies had reported that lncRNAs functioned as miRNA sponges to modulate neoplastic processes, we hypothesized that LINC00514 might act as a miRNA sponge in OS oncogenesis. The subcellular fraction assays clarified that LINC00514 was mainly located in cytoplasm (Figure 4A). With the aid of “miRDB” program, we found that miR-708, which had been verified to be a tumor suppressor in several cancer types, was a potential target of LINC00514. The complementary binding site between LINC00514 and miR-708 is presented in Figure 4B. In addition, the levels of miR-708 were determined by qPCR, indicating the lower expression of miR-708 in 107 OS tumor specimens (Figure 4C). Besides, qPCR analyses revealed that ectopic expression of LINC00514 remarkably decreased miR-708 levels, while LINC00514 deficiency notably elevated the levels of miR-708 in OS cells (Figure 4D). Vice versa, miR-708 overexpression significantly depressed LINC00514 levels, while miR-708 knockdown markedly increased LINC00514 levels (Figure 4E). Finally, we performed luciferase reporter assays. The results validated that when compared to that of the control group, the wild-type LINC00514/miR-708 mimics co-transfected group presented diminished luciferase activity, while no significant difference of luciferase activity was observed in the mutant-type LINC00514/miR-708 mimics co-transfected group (Figure 4F). Taken together, we discovered that miR-708 was a direct target of LINC00514 in OS cells.
SP1 Induced LINC00514 Expression by Acting as a Transcription Activator in OS Cells
Transcription factors (TFs) played crucial roles in modulating lncRNAs expression. To further investigate the regulator of LINC00514 in OS, we predicted the potential TFs which could bind to the promoter of LINC00514 using “Jaspar” program. The results showed that SP1, which was reported to activate diverse lncRNAs aberrant expression, was a potential TF which could promote LINC00514 expression (Figure 5A). Interestingly, SP1 was also upregulated in OS tumor samples (Figure 5B). To determine the regulatory effects of SP1 on LINC00514 expression, we silenced or overexpressed SP1 in 143B cells, and we discovered that LINC00514 was downregulated or upregulated in response to the knockdown or overexpression of SP1 in 143B cells (Figure 5C and D). In addition, we performed ChIP analyses to discover the exact binding site of SP1 in LINC00514 promoter and the result demonstrated that SP1 could bind to PS1 site of LINC00514 promoter (Figure 5E). Besides, luciferase reporter assays were conducted for further proving the binding relationship between SP1 and LINC00514 promoter, and we found that the effects of SP1 on the luciferase activity were remarkably promoted when PS1 site was wild-type, indicating that PS1 site of LINC00514 promoter was responsible for the binding of SP1 in OS cells (Figure 5F). Overall, SP1 transcriptionally activated LINC00514 and induced its expression in OS cells.
Discussion
The discovery of novel biomarkers for tumor screening is of great significance for the improvement of long-term survival of patients.24 Recently, increasing studies revealed lncRNAs as potential novel biomarkers due to their critical functions in epigenetic regulation as well as their frequent dysregulation in blood and tumor tissues of patients.25 In this study, a novel OS-related lncRNA, LINC00514, was identified by RT-PCR assays. Overexpression of LINC00514 was distinctly observed in both OS specimens and cell lines. Clinical study with 107 patients showed that patients with higher levels of LINC00514 had an advanced stage and positive metastasis, and exhibited a shorter overall survival. Moreover, LINC00514 was further demonstrated to be an independent poor prognostic factor for OS patients, which highlighted the great clinical values of LINC00514 as a novel prognostic biomarker. Previously, several lncRNAs were also reported to be frequently associated with advanced clinical stages and unfavorable prognosis of OS patients, such as lncRNA SNHG12 and lncRNA UCA1.26,27 Our findings, together with previous results, indicated that the positive associations between some functional lncRNA levels and patients’ prognosis may be a frequent event.
In cancer, the functional effects of lncRNAs acting as tumor promoters or oncogenes through transcriptional regulation of target genes have been frequently demonstrated in the abilities of tumor cells proliferation and metastasis.28,29 Our above results indicated that LINC00514 was highly expressed in OS, suggesting that it may act as a positive regulator in tumor progression. Thus, we performed a series of cellular experiments by silencing LINC00514 expression in 143B and MG63 using si-LINC00514. As expected, knockdown of LINC00514 distinctly inhibited tumor cell growth. Moreover, TUNEL assays demonstrated that knockdown of LINC00514 could suppress apoptosis of OS cells by increasing the activity of Caspase 3/9 which was confirmed by Western blot assays. In addition, we also provided evidence that LINC00514 silencing decreased OS cell migration and invasion. Overall, our findings suggested LINC00514 as an onco-lncRNA in OS.
The novel discovery of biological studies indicated that ceRNA which has emerged as a novel modulator in the posttranscriptional modification, which provided novel mechanisms involved in the regulation of gene expression.30,31 Several lncRNAs have been demonstrated to influence the tumorigenesis of various tumors via sponging tumor-related miRNAs.32,33 Thus, our group speculated that LINC00514 may display its carcinogenic roles by serving as a ceRNA. Firstly, LINC00514 was found to localize preferentially to the cytoplasm, indicating the possibility of LINC00514 as a mainly cytoplasmic lncRNA which can compete with ceRNAs to regulate miRNAs for further binding to their target mRNAs. Then, based on the data from bioinformatic assays and luciferase reporter assays, LINC00514 was confirmed to be a novel target of miR-708 which was an important tumor-related miRNA. Previously, dysregulation of miR-708 in several tumors and its potential roles in tumor progression had been frequently reported.34,35 In OS, miR-708 was found to be lowly expressed and suppress tumor cell proliferation and invasion by targeting URGCP.36 Hence, we also showed that the levels of miR-708 were upregulated in OS tissues. In addition, overexpression of LINC00514 could result in the suppression of miR-708 expressions. Thus, our findings, together with previous results, suggested that LINC00514 may promote the proliferation and metastasis of OS cells via sponging miR-708.
Transcriptional activation is an imperative mechanism leading to the overexpression of lncRNAs. Recently, several transcription factors have been reported to act as positive regulators in the modulation of lncRNAs expression in several tumors. For instance, lncRNA LINC00174 was shown to be upregulated in colorectal carcinoma and this upregulation was induced by STAT1 which was a transcription factor.21 Su et al37 reported that upregulation of lncRNA MIR100HG acting as a tumor promoter in OS was induced by the transcription factor ELK1. In addition, a common transcription factor, SP1, had been frequently reported to be involved in the regulation of the expression of lncRNAs and act as a promoter in may tumor progression.38,39 In this study, the results of bioinformatics assays predicted that SP1 could regulate LINC00514 transcription. Moreover, ChIP assays and Luciferase assays confirmed that SP1 could directly bind to LINC00514 promoter regions. These essential data suggested that SP1 activated LINC00514 translational expressions to increase LINC00514 in OS.
In conclusion, we firstly provided evidence that highly expressed LINC00514 acted as an oncogenic lncRNA that promoted the progression of OS through targeting miR-708. Upregulation of LINC00514 was associated with poor clinical prognosis and induced by SP1, target of miR-708. Our findings indicated that LINC00514 could be of interest in developing markers and therapeutic targets for OS patients.
Abbreviations
OS, Osteosarcoma; lncRNA, long non-coding RNA; CCK-8, Cell Counting Kit-8; qRT-PCR, quantitative real-time PCR; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; ChIP, Chromatin Immunoprecipitation; SDS-PAGE, dodecyl sulfate,sodium salt (SDS)-Polyacrylamide gel electrophoresis; PVDF, polyvinylidene fluoride. PCR, Polymerase Chain Reaction; ceRNA, Competing endogenous RNA; SP1, specificity protein 1; miRNA, micro RNA; PBS buffer, Phosphate-Buffered Saline buffer; TUNEL assay, Terminal deoxynucleotidyl transferase dUTP nick end labelling assay; siRNA, small interfering RNA; PS2, specificity protein 2; SNHG12, small nucleolar RNA host gene 12; UCA1, urothelial carcinoma-associated 1; URGCP, upregulator of cell proliferation; STAT1, signal transducer and activator of transcription 1; ELK1, ETS (E26 transformation-specific) Like-1 protein.
Ethics Approval
This study was conducted with permission by Ethics Review Committees of Zuanshiwan Hospital District of The Second Hospital of Dalian Medical University.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Ahmedin J. Cancer statistics, 2015. CA a Cancer J Clin. 2010;60(5):277–300. doi:10.3322/caac.20073
2. Vijayamurugan N, Bakhshi S. Review of management issues in relapsed osteosarcoma. Expert Rev Anticancer Ther. 2014;14(2):151–161. doi:10.1586/14737140.2014.863453
3. Klein MJ, Siegal GP. Osteosarcoma: anatomic and histologic variants. Am J Clin Pathol. 2006;125(4):555–581. doi:10.1309/UC6KQHLD9LV2KENN
4. Kumar R, Kumar M, Malhotra K, Patel S. Primary osteosarcoma in the elderly revisited: current concepts in diagnosis and treatment. Curr Oncol Rep. 2018;20(2):13. doi:10.1007/s11912-018-0658-1
5. Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14(11):722–735. doi:10.1038/nrc3838
6. Zamborsky R, Kokavec M, Harsanyi S, Danisovic L. Identification of prognostic and predictive osteosarcoma biomarkers. Med Sci (Basel, Switzerland). 2019;7:2.
7. Marino-Enriquez A, JV B. Molecular pathogenesis and diagnostic, prognostic and predictive molecular markers in sarcoma. Surg Pathol Clin. 2016;9(3):457–473. doi:10.1016/j.path.2016.04.009
8. Raimondi L, De Luca A, Costa V, et al. Circulating biomarkers in osteosarcoma: new translational tools for diagnosis and treatment. Oncotarget. 2017;8(59):100831–100851. doi:10.18632/oncotarget.19852
9. Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127(3):761–771. doi:10.1172/JCI84424
10. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi:10.1101/gad.1800909
11. Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi:10.1016/j.cell.2013.06.020
12. Akhade VS, Pal D, Kanduri C. Long noncoding RNA: genome organization and mechanism of action. Adv Exp Med Biol. 2017;1008:47–74.
13. Camacho CV, Choudhari R, Gadad SS. Long noncoding RNAs and cancer, an overview. Steroids. 2018;133:93–95. doi:10.1016/j.steroids.2017.12.012
14. Tang Y, Cheung BB, Atmadibrata B, et al. The regulatory role of long noncoding RNAs in cancer. Cancer Lett. 2017;391:12–19. doi:10.1016/j.canlet.2017.01.010
15. He R, Wu JX, Zhang Y, Che H, Yang L. LncRNA LINC00628 overexpression inhibits the growth and invasion through regulating PI3K/Akt signaling pathway in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(18):5857–5866. doi:10.26355/eurrev_201809_15915
16. Chen ZX, Chen CP, Zhang N, Wang TX. Low-expression of lncRNA FER1L4 might be a prognostic marker in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(8):2310–2314. doi:10.26355/eurrev_201804_14820
17. Tian ZZ, Guo XJ, Zhao YM, Fang Y. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. Int J Clin Exp Pathol. 2015;8(11):15138–15142.
18. Sun Q, Hao Q, Prasanth KV. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34(2):142–157. doi:10.1016/j.tig.2017.11.005
19. Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15(3):177–186. doi:10.1016/j.gpb.2016.12.005
20. Qi F, Liu X, Wu H, et al. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J Hematol Oncol. 2017;10(1):48. doi:10.1186/s13045-017-0420-4
21. Shen Y, Gao X, Tan W, Xu T. STAT1-mediated upregulation of lncRNA LINC00174 functions a ceRNA for miR-1910-3p to facilitate colorectal carcinoma progression through regulation of TAZ. Gene. 2018;666:64–71. doi:10.1016/j.gene.2018.05.001
22. Wang H, Huo X, Yang X-R, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. doi:10.1186/s12943-017-0680-1
23. Li X, Zhong W, Xu Y, Yu B, Liu H. Silencing of lncRNA LINC00514 inhibits the malignant behaviors of papillary thyroid cancer through miR-204-3p/CDC23 axis. Biochem Biophys Res Commun. 2019;508(4):1145–1148. doi:10.1016/j.bbrc.2018.12.051
24. Brown HK, Tellez-Gabriel M, Heymann D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017;386:189–195. doi:10.1016/j.canlet.2016.11.019
25. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
26. Zhou B, Li L, Li Y, Sun H, Zeng C. Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR-320a/MCL1 axis. Biomed Pharmacother. 2018;106:850–857. doi:10.1016/j.biopha.2018.07.003
27. Li W, Xie P, Ruan WH. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J Bone Oncol. 2016;5(2):80–85. doi:10.1016/j.jbo.2016.05.003
28. Thin KZ, Liu X, Feng X, Raveendran S, Tu JC. LncRNA-DANCR: a valuable cancer related long non-coding RNA for human cancers. Pathol Res Pract. 2018;214(6):801–805. doi:10.1016/j.prp.2018.04.003
29. Sun W, Yang Y, Xu C, Guo J. Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet. 2017;216–217:105–110. doi:10.1016/j.cancergen.2017.06.003
30. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
31. Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3(10):1113–1121. doi:10.1158/2159-8290.CD-13-0202
32. Li H, Zhang GY, Pan CH, Zhang XY, Su XY. LncRNA MAFG-AS1 promotes the aggressiveness of breast carcinoma through regulating miR-339-5p/MMP15. Eur Rev Med Pharmacol Sci. 2019;23(7):2838–2846. doi:10.26355/eurrev_201904_17561
33. Yang Q, Yu H, Yin Q, Hu X, Zhang C. lncRNA-NEF is downregulated in osteosarcoma and inhibits cancer cell migration and invasion by downregulating miRNA-21. Oncol Lett. 2019;17(6):5403–5408. doi:10.3892/ol.2019.10276
34. Liu B, Li R, Zhang J, et al. MicroRNA-708-3p as a potential therapeutic target via the ADAM17-GATA/STAT3 axis in idiopathic pulmonary fibrosis. Exp Mol Med. 2018;50(3):e465. doi:10.1038/emm.2017.311
35. Monteleone NJ, Lutz CS. miR-708-5p: a microRNA with emerging roles in cancer. Oncotarget. 2017;8(41):71292–71316. doi:10.18632/oncotarget.19772
36. Sui C, Liu D, Hu Y, Zhang L. MicroRNA-708-5p affects proliferation and invasion of osteosarcoma cells by targeting URGCP. Exp Ther Med. 2019;17(3):2235–2241. doi:10.3892/etm.2019.7171
37. Su X, Teng J, Jin G, et al. ELK1-induced upregulation of long non-coding RNA MIR100HG predicts poor prognosis and promotes the progression of osteosarcoma by epigenetically silencing LATS1 and LATS2. Biomed Pharmacother. 2019;109:788–797. doi:10.1016/j.biopha.2018.10.029
38. Hu X-H, Dai J, Shang H-L, Zhao Z-X, Hao Y-D. SP1-mediated upregulation of lncRNA ILF3-AS1 functions a ceRNA for miR-212 to contribute to osteosarcoma progression via modulation of SOX5. Biochem Biophys Res Commun. 2019;511(3):510–517. doi:10.1016/j.bbrc.2019.02.110
39. Silva G, Marins M, Fachin AL, Lee SH, Baek SJ. Anti-cancer activity of trans-chalcone in osteosarcoma: involvement of Sp1 and p53. Mol Carcinog. 2016;55(10):1438–1448. doi:10.1002/mc.22386
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.