Back to Journals » OncoTargets and Therapy » Volume 12
SOX30 methylation correlates with disease progression in patients with chronic myeloid leukemia
Authors Zhang TJ, Wen XM, Zhou JD , Gu Y, Xu ZJ, Guo H, Ma JC, Yuan Q, Chen Q, Lin J, Qian J
Received 27 March 2019
Accepted for publication 28 May 2019
Published 20 June 2019 Volume 2019:12 Pages 4789—4794
DOI https://doi.org/10.2147/OTT.S210168
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Ting-Juan Zhang,1–3,* Xiang-Mei Wen,2–4,* Jing-Dong Zhou,1–3,* Yu Gu,1–3 Zi-Jun Xu,2–4 Hong Guo,2–4 Ji-Chun Ma,2–4 Qian Yuan,1–3 Qin Chen,2–4 Jiang Lin,2–4 Jun Qian1–3
1Department of Hematology, Affiliated People’s Hospital of Jiangsu University, Zhenjiang, Jiangsu, People’s Republic of China; 2Zhenjiang Clinical Research Center of Hematology, Zhenjiang, Jiangsu, People’s Republic of China; 3The Key Lab of Precision Diagnosis and Treatment of Zhenjiang City, Zhenjiang, Jiangsu, People’s Republic of China; 4Laboratory Center, Affiliated People’s Hospital of Jiangsu University, Zhenjiang, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Background: Our previous study has reported that aberrant SOX30 methylation was associated with poor prognosis in AML, and it correlated with disease progression in MDS. Herein, we further determined SOX30 methylation and its clinical significance in the other myeloid malignance – chronic myeloid leukemia (CML).
Methods: SOX30 methylation was examined by real-time quantitative methylation-specific PCR and bisulfite sequencing PCR, whereas SOX30 expression was detected by real-time quantitative PCR.
Results: SOX30 methylation was identified in 11% (10/95) CML patients. SOX30 methylation was associated with lower hemoglobin and platelets (P=0.006 and 0.032, respectively). Importantly, significant differences were observed in the distributions of clinical stages and cytogenetics (P=0.006 and 0.002, respectively). The frequency of SOX30 methylation in chronic phase (CP) stage occurred with lowest frequency (4/74, 5%), higher in accelerated phase (AP) stage (1/7, 14%), and the highest in blast crisis (BC) stage (12/31, 39%). In addition, SOX30 methylated patients tended to have a higher level of BCR-ABL transcript than SOX30 non-methylated patients (P=0.063). In two paired CML patients, SOX30 methylation showed lower density in CP stage (19% and 17%, respectively), and was significantly increased in BC stage (89% and 69%, respectively) during disease progression. Additionally, SOX30 methylated CML patients presented a lower SOX30 transcript level than SOX30 non-methylated CML patients (P=0.046).
Conclusion: Our study revealed that SOX30 methylation correlated with disease progression in chronic myeloid leukemia.
Keywords: SOX30, methylation, chronic myeloid leukemia, progression
Introduction
Chronic myeloid leukemia (CML) was the first human cancer to be associated with a consistent chromosomal abnormality, namely the Philadelphia (Ph) chromosome, which generates from a reciprocal translocation between chromosome 9 and 22.1 In its natural history, CML is usually diagnosed in chronic phase (CP) and then progresses through an accelerated phase (AP) to a nearly invariably fatal blast crisis (BC).1 Without effective treatment, CML patients in CP progress to advanced stages, the prognosis for which is poor.1 However, until now, the underlying mechanisms of CML progression are varied and not entirely understood. So far the best characterized includes the accumulation of molecular and chromosomal abnormalities.2,3 Recently, aberrant DNA methylation through the activation of tumor suppressor genes playing a role in the progression of CML has been aroused great attention.4
SOX30, a relatively new member of the SOX family, encodes transcription factor belonging to the high mobility group (HMG) superfamily.5 It has been considered to be involved in spermatogonial differentiation and spermatogenesis.6 For years, the potential role of SOX30 in human cancers remains poorly defined. Recently, SOX30 has been identified as a key tumor suppressor gene mediated by promoter methylation in tumorigenesis including lung cancer, acute myeloid leukemia (AML), and myelodysplastic syndromes (MDS).7,8 In clinics, SOX30 had a favorable prognostic impact on lung adenocarcinoma patients.9 Moreover, aberrant SOX30 methylation was associated with poor prognosis in AML, whereas it correlated with disease progression in MDS.8 In this study, we further determined SOX30 methylation and its clinical significance in CML.
Patients and methods
Patients and samples
The study was approved by the Ethics Committee and Institutional Review Board of the Affiliated People’s Hospital of Jiangsu University in accordance with the Declaration of Helsinki. After signing the written informed consents, bone marrow (BM) was collected from 95 CML patients at diagnosed time as well as 28 healthy donors seen as controls. The main clinical and laboratory features of CML patients were presented in Table 1. Treatment of CML patients at CP stage received tyrosine kinase inhibitors (TKI)-based therapy, whereas CML patients at AP/BC stage received chemotherapy together with TKI-based therapy. BM mononuclear cells (BMMNCs) were separated by density-gradient centrifugation using Lymphocyte Separation Medium (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). When karyotypes were analyzed, BM cells were harvested after 1 to 3 days of unstimulated culture in RPMI 1640 medium containing 20% fetal calf serum (ExCell Bio, Shanghai, China).
 | Table 1 Comparison of clinical and laboratory features between CML patients with and without SOX30 methylation |
RNA isolation, reverse transcription and RQ-PCR
Total RNA isolation and reverse transcription were as reported previously.10 The expression of SOX30 was detected by real-time quantitative PCR (RQ-PCR) as reported previously.8 The housekeeping gene ABL detected by RQ-PCR using 2×SYBR Green PCR Mix (Multisciences, Hangzhou, China) was used to calculate the abundance of SOX30 transcript. Relative SOX30 transcript level was calculated by 2−∆∆CT methods.
DNA isolation, bisulfite modification, and RQ-MSP
Genomic DNA isolation and modification were performed as reported previously.10 The methylation level of SOX30 was examined by real-time quantitative methylation-specific PCR (RQ-MSP) using AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ, USA) as reported previously.8 Relative SOX30 methylation level was calculated by 2−∆∆CT methods.
BSP
Bisulfite sequencing PCR (BSP) and clone sequencing were conducted as reported previously.8 Six independent clones of BSP products were sequenced.
Statistical analysis
SPSS software version 20.0 and GraphPad Prism 5.0 were applied to perform statistical analysis. Mann-Whitney’s U test was carried to compare the difference of continuous variables between two groups, whereas Pearson Chi-square analysis/Fisher exact test was applied to compare the difference of categorical variables between two groups. Correlation analysis was performed by Spearman test. All tests were two sided, and P<0.05 was defined as statistically significant.
Results
Correlation of SOX30 methylation with clinical/laboratory characteristics in CML
Our previous study has reported aberrant SOX30 methylation in AML and MDS, and SOX30 hypermethylation correlated with poor prognosis in AML, also associated with disease progression in MDS.8 Herein, we further determined SOX30 methylation in the other myeloid malignance CML using RQ-MSP. According to the previously set cut-off value of 1.024, SOX30 methylation was identified in 11% (10/95) CML patients, and was a bit higher than controls (0%, 0/28) (Figure 1, P=0.115). In order to analyze the clinical significance in CML, we further compared the clinical and laboratory features between SOX30 methylated and non-methylated patients (Table 1). No significant differences was shown in age and white blood cells (P>0.05). However, SOX30 methylation was associated with lower hemoglobin and platelets (P=0.006 and 0.032, respectively). Importantly, significant differences were observed in the distributions of clinical stages and cytogenetics (P=0.006 and 0.002, respectively). Among different clinical stages, the frequency of SOX30 methylation in chronic phase (CP) stage occurred with lowest frequency (4/74, 5%), higher in accelerated phase (AP) stage (1/7, 14%), and the highest in blast crisis (BC) stage (12/31, 39%). For cytogenetics, t(9;22) with additional alterations patients showed the highest incidence of SOX30 methylation (4/11, 36%), whereas cases with t(9;22) patients presented the lowest incidence of SOX30 methylation (2/63, 3%). In addition, SOX30 methylated patients tended to have a higher level of BCR-ABL transcript than SOX30 non-methylated patients (P=0.063). All the positive results indicated that SOX30 methylation was associated increased clinical stage and may play a role in disease progression in CML.
SOX30 methylation alterations during disease progression in paired CML patients
To confirm whether SOX30 was associated with CML progression, we next determined SOX30 methylation at different clinical stages of two paired CML patients during disease progression. Detected by BSP, both two CML patients showed low density of SOX30 methylation in CP stage (19% and 17%, respectively), and the density of SOX30 methylation was significantly increased in BC stage during disease progression (89% and 69%, respectively) (Figure 2).
Association of SOX30 methylation with its expression in CML
As is well known, DNA methylation in promoter-associated CpG islands plays a direct role in gene silencing that is one of the drivers of neoplastic transformation through the inactivation of critical tumor-suppressor pathways. We further determined SOX30 transcript level in 52 CML patients with available mRNA samples. Although we did not observe the significantly negative association between SOX30 methylation and SOX30 expression (R=−0.169, P=0.232, n=52, Spearman test), SOX30 methylated CML patients presented a lower SOX30 transcript level than SOX30 non-methylated CML patients (P=0.046, Figure 3).
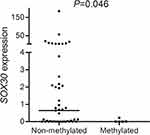 | Figure 3 SOX30 expression level in SOX30 methylated and non-methylated CML patients. SOX30 methylation level and expression level in 52 CML patients were examined by RQ-MSP and RQ-PCR, respectively. |
Discussion
Following our previous study, herein, we further for the first time reported SOX30 methylation in CML. Although it was not a frequent event in CML, SOX30 methylation was associated with advanced clinical stage, and may have a crucial role in CML progression. The mechanisms of CML progression are varied and not entirely understood. So far the best characterized includes differentiation arrest, genomic instability, telomere shortening and loss of tumor-suppressor functions.2 Mechanically, cytogenetic abnormalities and molecular alterations, such as double Ph chromosome, trisomy chromosome 8, i(17q), trisomy chromosome 19, t(3;21)(q26;q22), t(7;11)(p15;p15), p53 mutations, RAS mutation, and increased BCR-ABL transcript, are pathogenetically linked to the progression of CML.3 Recently, epigenetic changes especially aberrant DNA methylation associated with gene silencing are also identified to play crucial roles in the disease progression of CML.4 For instance, SHP-1 hypermethylation was involved in the progression in CML through dysregulating BCR-ABL1, AKT, MAPK, MYC and JAK2/STAT5 signaling pathways.11 In addition, our previous studies have revealed that hypermethylation of ID4 and DLX4 was related to disease progression in CML, and ID4 also had a direct role in affecting cell proliferation and apoptosis in K562 cell-line.12,13
Although we have proved that SOX30 methylation was associated with CML progression in clinics, the direct role of SOX30 in the pathogenesis of CML was not studied. Recently, SOX30 has been validated to be a tumor suppressor gene in several solid cancers with diagnostic and prognostic value. Han et al demonstrated that SOX30 was silenced caused by promoter methylation and functioned as a novel tumor suppressor partly by transcriptional activating p53 or activating the transcription of desmosomal genes in lung cancer.7,14 In addition, SOX30 could also inhibited tumor metastasis through attenuating Wnt signaling via the regulation of β-Catenin in a transcriptional and posttranslational manner in lung cancer.15 In clinics, the expression of SOX30 was verified to be closely associated with clinical outcomes in lung cancer patients.9 Moreover, SOX30 expression was identified as prognostic biomarkers in several other human cancers such as bladder cancer and advanced-stage ovarian cancer.16,17 In colon cancer and hepatocellular carcinoma, the anti-cancer effects of SOX30 rescued the tumor-promoting effect mediated by miR-645 overexpression.18,19 However, no functional studies showed the role of SOX30 in hematological malignances. Moreover, the current study could not further determine the impact of SOX30 methylation in CML due to limited cases with survival data. Obviously, further studies are needed to determine the direct role of SOX30 during CML progression and its clinical implication.
In summary, SOX30 methylation correlated with disease progression in CML, and provided novel insights into CML biology acting as a potential therapeutic target against disease progression.
Ethics statements
This study approved by the Ethics Committee and Institutional Review Board of the Affiliated People’s Hospital of Jiangsu University in accordance with the Declaration of Helsinki.
Acknowledgments
This study was supported by National Natural Science Foundation of China (81270630), Medical Innovation Team of Jiangsu Province (CXTDB2017002), 333 Project of Jiangsu Province (BRA2016131), Six Talent Peaks Project in Jiangsu Province (2015-WSN-115), Zhenjiang Clinical Research Center of Hematology (SS2018009), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_2281, KYCX17_1821), Social Development Foundation of Zhenjiang (SH2017040, SH2018044), Clinical Medical Science Development Foundation of Jiangsu University (JLY20160011), Natural Science Foundation of Jiangsu Province for Youths (BK20180280) and Scientific Research Foundation of Affiliated People’s Hospital of Jiangsu University for Ph.D. (KFB201603).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Apperley JF. Chronic myeloid leukaemia. Lancet. 2015;385(9976):1447–1459. doi:10.1016/S0140-6736(13)62120-0
2. Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7(6):441–453. doi:10.1038/nrc2147
3. Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103(11):4010–4022. doi:10.1182/blood-2003-12-4111
4. Heller G, Topakian T, Altenberger C, et al. Next-generation sequencing identifies major DNA methylation changes during progression of Ph+ chronic myeloid leukemia. Leukemia. 2016;30(9):1861–1868. doi:10.1038/leu.2016.143
5. Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227(2):239–255. doi:10.1006/dbio.2000.9883
6. Osaki E, Nishina Y, Inazawa J, et al. Identification of a novel Sry-related gene and its germ cell-specific expression. Nucleic Acids Res. 1999;27(12):2503–2510. doi:10.1093/nar/27.12.2503
7. Han F, Liu W, Jiang X, et al. SOX30, a novel epigenetic silenced tumor suppressor, promotes tumor cell apoptosis by transcriptional activating p53 in lung cancer. Oncogene. 2015;34(33):4391–4402. doi:10.1038/onc.2014.370
8. Zhou JD, Wang YX, Zhang TJ, et al. Identification and validation of SRY-box containing gene family member SOX30 methylation as a prognostic and predictive biomarker in myeloid malignancies. Clin Epigenetics. 2018;10:92.
9. Han F, Liu W, Xiao H, et al. High expression of SOX30 is associated with favorable survival in human lung adenocarcinoma. Sci Rep. 2015;5:13630. doi:10.1038/srep13630
10. Zhang TJ, Zhou JD, Zhang W, et al. H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin Epigenetics. 2018;10:47. doi:10.1186/s13148-018-0486-z
11. Li Y, Yang L, Pan Y, Yang J, Shang Y, Luo J. Methylation and decreased expression of SHP-1 are related to disease progression in chronic myelogenous leukemia. Oncol Rep. 2014;31(5):2438–2446. doi:10.3892/or.2014.3098
12. Zhou JD, Zhang TJ, Li XX, et al. Epigenetic dysregulation of ID4 predicts disease progression and treatment outcome in myeloid malignancies. J Cell Mol Med. 2017;21(8):1468–1481. doi:10.1111/jcmm.13073
13. Zhou JD, Wang YX, Zhang TJ, et al. Epigenetic inactivation of DLX4 is associated with disease progression in chronic myeloid leukemia. Biochem Biophys Res Commun. 2015;463(4):1250–1256. doi:10.1016/j.bbrc.2015.06.095
14. Hao X, Han F, Ma B, et al. SOX30 is a key regulator of desmosomal gene suppressing tumor growth and metastasis in lung adenocarcinoma. J Exp Clin Cancer Res. 2018;37(1):111. doi:10.1186/s13046-018-0778-3
15. Han F, Liu WB, Shi XY, et al. SOX30 inhibits tumor metastasis through attenuating Wnt-signaling via transcriptional and posttranslational regulation of β-catenin in lung cancer. EBioMedicine. 2018;31:253–266. doi:10.1016/j.ebiom.2018.04.026
16. Liu Y, Wang H, Zhong J, et al. Decreased expression of SRY-box containing gene 30 is related to malignant phenotypes of human bladder cancer and correlates with poor prognosis. BMC Cancer. 2018;18(1):642. doi:10.1186/s12885-018-4242-8
17. Han F, Liu WB, Li JJ, et al. SOX30 is a prognostic biomarker and chemotherapeutic indicator for advanced-stage ovarian cancer. Endocr Relat Cancer. 2019;303–319. doi:10.1530/ERC-18-0529
18. Guo ST, Guo XY, Wang J, et al. MicroRNA-645 is an oncogenic regulator in colon cancer. Oncogenesis. 2017;6(5):e335. doi:10.1038/oncsis.2017.37
19. Tao J, Liu Z, Wang Y, et al. MicroRNA-645 represses hepatocellular carcinoma progression by inhibiting SOX30-mediated p53 transcriptional activation. Int J Biol Macromol. 2019;121:214–222. doi:10.1016/j.ijbiomac.2018.10.032
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


