Back to Journals » Cancer Management and Research » Volume 13
SOX10 Knockdown Inhibits Melanoma Cell Proliferation via Notch Signaling Pathway
Received 14 July 2021
Accepted for publication 19 August 2021
Published 17 September 2021 Volume 2021:13 Pages 7225—7234
DOI https://doi.org/10.2147/CMAR.S329331
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Youqun Tang, Yanming Cao
Department of Oncology, The Third Xiangya Hospital, Central South University, Changsha, 410013, Hunan, People’s Republic of China
Correspondence: Yanming Cao
Department of Oncology, The Third Xiangya Hospital, Central South University, Changsha, 410013, Hunan, People’s Republic of China
Email [email protected]; [email protected]
Purpose: Melanoma is a serious and malignant disease worldwide. Seeking diagnostic markers and potential therapeutic targets is urgent for melanoma treatment. SOX10, a member of the SoxE family of genes, is a transcription factor which can regulate the transcription of a wide variety of genes in multiple cellular processes.
Methods: The mRNA level and protein expression of SOX10 is confirmed by bioinformatic analysis and IHC staining. MTT, clone formation and EdU analysis showed that SOX10 knockdown (KD) could significantly inhibit melanoma cell proliferation. FACS analysis showed that SOX10 KD could markedly enhance the level of cell apoptosis. The downstream target signaling pathway is predicted by RNA-seq based on the public GEO database. The activation of Notch signaling mediated by SOX10 is tested by qPCR and Western blot.
Results: Ectopic upregulation of SOX10 was found in melanoma patient tissues compared to normal nevus tissues in mRNA and protein levels. Furthermore, both mRNA and protein level of SOX10 were negatively correlated with melanoma patient’s prognosis. SOX10 knockdown could obviously suppress the proliferation ability of melanoma cells by inactivating Notch signaling pathway.
Conclusion: Our study confirmed that SOX10 is an oncogene and activate Notch signaling pathway, which suggests the potential treatment for melanoma patients by target SOX10/Notch axis.
Keywords: SOX10, melanoma, bioinformatics, proliferation, biomarkers
Introduction
Melanoma is a type of aggressive malignant neoplasm that originates from the skin and mucosa with high mortality rates.1 With the development of medical science, melanoma treatment has entered a phase of diversified treatment. However, the melanoma treatment remains a challenge because of its rarity and heterogeneity.2 Therefore, it is an urgent need to look for novel therapeutic targets for melanoma.
Sex determining region Y-box 10 (SOX10), a member of the SoxE family of genes, play a key role in the embryonic development of melanocytes. This gene is highly expressed in premigratory neural crest cells, which can regulate proliferation, survival, and differentiation ability of peripheral glial cells and pigment cells.3 SOX10 deficiency can induce embryonic lethality, aganglionosis of the colon and pigmentation defects in mice.4 SOX10 significantly regulates cancer proliferation, migration and apoptosis, and is closely related to tumor progression.5 Furthermore, SOX10 can increase the activity of dopachrome tautomerase and tyrosinase in melanocyte lineage.6
Only recently, many studies indicated that SOX10 was involved in the formation, development and progression of melanoma. Previous studies suggesting that the frequencies of SOX10 DNA alteration was extremely low in melanoma patients, indicating that SOX10 wild type was mainly maintain the melanoma development and progression.7 Different mouse model studies indicated that SOX10, as an important regulator, accelerate melanomagenesis.8,9 Graf et al found that SOX10 can control melanoma inhibitory activity to accelerate melanoma cell metastasis.10 Recently, another study indicated that SOX10 can regulate myelin protein PMP2 to promote the migration and invasion of melanoma cell.11 However, the molecular mechanism of how SOX10 acts is unclear in melanomagenesis.
In our study, we investigated the expression of SOX10 in melanoma based on bioinformatic analysis and IHC staining. We also knock down the SOX10 expression in melanoma cell, and confirmed the effect of SOX10 knockdown (KD) on the proliferation ability in melanoma cell. We used the GEO and PDB database to explore the molecular mechanism. Moreover, we validated SOX10 can regulate Notch signaling pathway to accelerate melanomagenesis, which seems to be responsible for the proliferation promoter of SOX10.
Materials and Methods
Bioinformatic Analysis
Bioinformatic analysis was performed on the GSE37059 dataset (https://www.ncbi.nlm.nih.gov/gds/?term=) that was submitted by Shakhova et al.8 Then, this data was analyzed by R packages (pheatmap and ggplot2) to get volcano plots and heat map. The Gene Ontology (GO) enrichment and KEGG enrichment analysis was based on the DAVID database (https://david.ncifcrf.gov/).12 The expression profiles of SOX10 among 33 cancer types were based on TCGA database (https://www.cancer.gov/).13 The tool, GEPIA (http://gepia.cancer-pku.cn/), was used for data visualization.14 Protein Data Bank (PDB) database (https://www.rcsb.org/) was utilized to visualize the protein structure of SOX10.15 The single-cell sequencing analysis and IHC staining analysis was based on HPA database (https://www.proteinatlas.org/).16
Cell Culture
The melanoma cell lines (A375 and HT144) were obtained from the American Type Culture Collection (ATCC), culturing in Dulbecco’s Modified Eagle Medium (DMEM) medium (Cat no. SH30022.01, Hyclone) with 10% FBS (Cat no. 10091-148, Gibco), 100U/mL penicillin (Cat no. P1400, Solarbio) at 37°C in humidified incubator in the presence of 5%CO2.
shRNA Transfection
For knockdown experiments pSuper-Neo-GFP plasmids (Oligoengine) were used that expressed a Sox10-specific shRNA (targeted region: 5′-CTGCTGTTCCTTCTTGACCTTGCCC-3′) or a scrambled control shRNA. A375 and HT144 cells seeded at 2.5×105 cells per well in a 6-well plate transfected with 12 μg of plasmid DNA for 5 hours, respectively, using Lipofectamine™ 2000 (Life Technologies, USA). After 2 days of cell culture, the knockdown efficiency was confirmed by Western blot analysis. Scramble-shRNA transfected cells were used as the negative control for SOX10-knockdown cells.
Clinical Samples
A whole of 54 samples includes 40 melanoma tissues and 14 normal nevus tissue samples (paraffin embedded section) were surgically resected at The Third Xiangya Hospital of Central South University (China). The collection and use of tissues were performed according to the ethical standards formulated in the Declaration of Helsinki. Written informed consent was obtained from each patient, and the study was approved by the research ethics committee of the Third Xiangya Hospital of Central South University. Patients did not receive any preoperative chemotherapy or chemoradiotherapy. The final immunohistochemistry scores were blindly scored by two pathologists.
IHC Staining
The IHC staining was determined by using immunohistochemistry kit (Maixin Biotech. Co., Fuz-hou, China) following the manufacturer’s procedures. The specific methods were as follows: the sections of melanoma and normal nevus tissue was deparaffinized by standard xylene-deparaffin procedure, which were subsequently dehydrated with ethanol. Antigen retrieval was carried out by citric acid and a pressure cooker. Then, enzyme closure was performed using with 3% hydrogen peroxide. These sections were incubated overnight at 4°C with SOX10 primary antibodies (ab227684, Abcam). Immunohistochemistry without the primary antibody was used as a negative control. After incubation with anti-SOX10, these sections were then washed by PBST and incubated with secondary antibodies. Finally, staining was visualized by DAB and counterstaining was performed with haematoxylin.
MTT Analysis
Transient transfected HT144 and A375 cells were plated in 96-well plates (5x103 cells/well) and cultured for 24, 48 and 72 hours at 37°C. Cell viabilities were then evaluated with a MTT kit (5 mg/mL, Sigma‑Aldrich; Merck KGaA) at 24, 48 and 72 hours, respectively. Then, 150 µL DMSO was added to dissolve the precipitates and the effect of cell number on absorbance at 490 nm was measured using a microplate reader (Molecular Devices LLC).
Clone Formation Assay
Transient transfected HT144 and A375 cells were plated in 24-well plates (100 cells/well), which were selected using 5 mg/mL puromycin. Two weeks later, the surviving colonies on the 24-well plates were fixed with 4% paraformaldehyde and stained with 1% crystal violet.
EdU Analysis
EdU, a thymine nucleoside analogue, can replace thymine (T) to infiltrate replicating DNA molecules during cell proliferation, and then detect cell proliferation by immunofluorescence technique and specific reaction with Apollo fluorescent dye respectively. This is a new non-radioisotope cell proliferation assay. Transient transfected HT144 and A375 cells were plated in 96-well plates. The activity of DNA replication was determined by using Edu cell proliferation kit (RiboBio, Guangzhou, China) following the manufacturer’s procedures. Finally, the results were captured under a fluorescence microscope.
Apoptosis Analysis
The above treated cells were digested and collected with trypsin without EDTA. The cells were washed with PBS twice, centrifuged at 2000rpm for 5 min each time, and about 3.2×105 cells were collected. Then, 500ul Binding Buffer was added to suspend cells, which were add 5ul Annexin V-APC (KeyGEN BioTECH, JiangSu, China) and add 5ul Propidium Iodide and mix well. The reaction was observed by flow cytometry after 10 min.
Quantitative Real-Time PCR
RNA was extracted with an RNA isolation kit (Tiangen, Beijing, China). A reverse transcription kit (TaKaRa Biotechnology) was used to perform reverse transcription. Quantitative real‐time PCR (qRT‐PCR) was performed using a quantitative SYBR Green PCR Kit (Takara Bio). Primers used were listed as follows: NOTCH1 forward: TGAATGGCGGGAAGTGTGAAG; NOTCH1 reverse: GGTTGGGGTCCTGGCATCG; GAPDH forward: GTCTCCTCTGACTTCAACAGCG, GAPDH reverse: ACCACCCTGTTGCTGTAGCCAA.
Western Blot
Western blot assays were performed as previously described.17 In brief, total proteins from melanoma cells were extracted in RIPA buffer (Sigma-Aldrich; Merck KGaA) on ice (the lysis buffer contain phosphatase and protease inhibitors), Then, the lysates were prepared with lysis buffer (Beyotime), and the protein concentration was determined via the BCA assay. Approximately 50ug of protein was loaded into a 12% SDS/Polyacrylamide gel. Electrophoresis was carried out at 80 V for 20 min followed by 40 min at 120 V. Western blot analysis was performed following protein transfer to a PVDF membrane. After blocking with 5% nonfat milk, the membranes were incubated at 4 °C overnight with SOX10 (ab227684, Abcam), Notch1 (ab52627, Abcam), NICD (ab52301, Abcam), Hes1 (ab108937, Abcam), and GAPDH (ab8245, Abcam) antibody. Membranes were visualised with Electrochemiluminescence Plus Detection Reagents.
Statistical Analysis
Data were analyzed using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA) and presented as mean standard deviation (SD). Two-tailed Student’s t-tests were used to determine p values.
Results
The Expression of SOX10 in Melanoma Based on Bioinformatic Analysis and IHC Staining
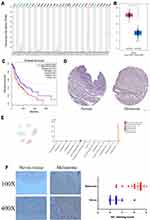
Firstly, we extracted the data files of SOX10 in multiple cancer types based on TCGA database. The mRNA level of SOX10 is decreased in bladder urothelial carcinoma, breast invasive carcinoma, cervical squamous cell carcinoma and endocervical adenocarcinoma, colon adenocarcinoma, esophageal carcinoma, pancreatic adenocarcinoma, rectum adenocarcinoma, testicular germ cell tumors, but only increased in melanoma significantly (Figure 1A and B). Moreover, we also found the SOX10 mRNA level was significantly and obviously negatively correlated with prognosis in melanoma patients (Figure 1C). Moreover, we utilized HPA database to confirm the SOX10 protein expression in melanoma tissues and normal nevus tissues, which indicated the expression of SOX10, primarily located in cell nuclear, was obviously increased in melanoma patients compared to normal nevus tissues (Figure 1D). Single cell sequencing analysis indicated that SOX10 was primarily expressed in pigment cells, and slightly expressed in macrophages, fibroblasts, basal keratinocytes, suprabasal keratinocytes, smooth muscle cells, and endothelial cells (Figure 1E). IHC staining indicated that SOX10 expression is obviously enhanced in melanoma patients compared to normal nevus tissues (Figure 1F).
SOX10 Knockdown Can Markedly Reduce the Proliferation Ability and Increase the Apoptosis Level of Melanoma Cells
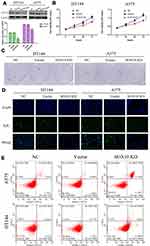
To further investigate the biological function of SOX10 in melanoma cells. We inhibited the expression of SOX10 in melanoma cells, HT144 and A375, via SOX10 shRNA (Figure 2A). Then, the MTT analysis was used to confirmed the effect of SOX10 KD on the proliferation ability of HT144 and A375 cells (Figure 2B), which indicated that SOX10 inhibiting could significantly repress the melanoma cell viability. The clone formation assay also indicated that SOX10 deficiency could attenuate HT144 and A375 cell proliferation (Figure 2C). Furthermore, EdU analysis showed that the DNA replication level was obviously decreased in SOX10 KD group compared to NC and Vector group (Figure 2D), which indicated that SOX10 KD repress the proliferation ability via reducing DNA replication in melanoma cell. Moreover, we found SOX10 KD in melanoma cell could obviously increase the apoptosis level (Figure 2E), suggesting that SOX10 could accelerate the development and progression of melanoma via inhibiting cell apoptosis.
The GO Function and Enriched Pathways of SOX10 KD in Melanoma Based on Bioinformatics
To dissect the molecular mechanism of SOX10 in melanoma, we extracted the GSE37059 datasets in GEO database.8 The volcano plots showed that there were 115 upregulated genes and 290 downregulated genes when SOX10 KD (Figure 3A and B). We utilized GO and KEGG enrichment analysis to predicted potential biological functions and molecular mechanism of these key genes. In the terms of KEGG, we found the upregulated genes were primarily distributed to Notch signaling pathway, DNA replication, and cell cycle, the downregulated genes were markedly enriched in TNF signaling pathway, NF-kappa B signaling pathway, and cytosolic DNA-sensing pathway (Figure 3C). In the terms of GO, we found these upregulated genes were obviously distributed in pigmentation, melanocyte differentiation, melanin biosynthetic process, and meiotic cell cycle, these downregulated genes were markedly enriched in response to virus, response to tumor necrosis factor, and regulation of endopeptidase activity (Figure 3D). Therefore, these results indicated SOX10 might regulate Notch signaling pathway to accelerate DNA replication and cell cycle in melanomagenesis.
SOX10 KD Decelerate Melanoma Cells Proliferation by Inactivating Notch Signaling
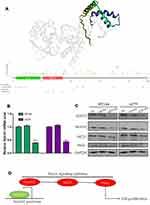
In the results of the last section, we found SOX10 might activate Notch pathway, but the specific molecular mechanism remains unclear. Therefore, we confirmed the protein secondary and tertiary structure based on PDB database, which showed that SOX10 protein has SOX_N and HMG_box domain that could bind to DNA activate transcription (Figure 4A). These HMG_box domain indicated that SOX10 might bind to Notch1 DNA promoter to increase the Notch1 mRNA level. Therefore, we detected the mRNA level of Notch1 in HT144 and A375 cell lines, which indicated that SOX10 KD could significantly repress the Notch1 mRNA level compared to NC and vector group (Figure 4B). We further confirmed the effect of SOX10 on Notch1 signaling pathway by Western blot, suggesting that SOX10 KD could obviously reduce the expression of Notch signaling pathway protein such as, Notch1, NICD, and Hes1 (Figure 4C).
Discussion
In our study, we found SOX10 mRNA and protein levels were both significantly increased in melanoma patients. The high mRNA and protein levels of SOX10 can both indicate the poor prognosis for melanoma patients, which suggests that SOX10 might be an excellent biomarker in melanoma. Our data also demonstrate the previously role of SOX10 in promoting melanoma cell proliferation at later time points.18 Furthermore, we validated Notch1 as a direct downstream gene for SOX10, resulting in activating Notch signaling pathway in melanoma.
SOX10 has been indicated that it could be a sensitive biomarker for melanoma patients based on IHC staining.19–21 Moreover, the expression of SOX10 was also significantly enhanced in multiple melanoma cell lines, such as SbCI2, WM3211, WM35, WM278, WM793, WM293A, WM9, WM1232, 451Lu and 1205Lu.10 In our study, we found SOX10 mRNA and protein were both enhanced in melanoma patients. Moreover, both SOX10 mRNA and protein level were negatively correlated with melanoma patient’s prognosis. Graf et al found that SOX10 could promote melanoma cell invasion by regulating melanoma inhibitory activity.10 Shakhova et al also found that SOX10 could promote the formation and maintenance of giant congenital naevi and melanoma.8 Loftus SK. Also indicated that inhibiting SOX10 could markedly reduce the proliferation ability in melanoma.22 Similarly, in our study, we found that SOX10 KD could suppress the melanoma cell proliferation.
The effect of SOX10 on melanoma cell proliferation has been well demonstrated. However, the further downstream mechanism of SOX10 is still unclear. Bioinformatics was useful for the biologist to investigate the potential mechanism and interrelation of molecules.23 Shakhova et al utilized RNA-seq analysis to explore the downstream molecular mechanism of SOX10 in melanoma.8 Therefore, we also used this datasets (GSE37059) from GEO database. We found SOX10 KD in melanoma can induce multiple signaling pathway alterations based on the GSE37059 dataset, especially in Notch signaling pathway.
SOX10, as an important transcription factor with 466 amino acids, is essential for the cell survival, and maintenance of pluripotency. The gene sox10 is located on chromosome 22q13.1.24 But the alteration of sox10 gene is very scarce in melanoma, which indicated that the wild type SOX10 function is needed for the formation, development and progression of melanoma.7 Therefore, we analyzed the secondary and tertiary protein structure of wild type SOX10. Our data indicated that wild type of SOX10 still has a transcriptional regulatory function. Therefore, we confirmed the mRNA level of North 1 was decreased by SOX10 knockdown.
It is well known that the Notch signalling pathway is involved in the progression of epidermal homeostasis. Moreover, ectopically activation can promote the development and progression of many cancers.25,26 Notch signaling pathway is involved in many physiologic processes, such as proliferation, apoptosis, migration, invasion and tumorigenesis, either as a tumor promoter or suppressor depending on the cellular context, level of expression and cross-talk with other signaling systems.27 Howard et al found that activating Notch3 in melanoma cell lines leads to enhanced cancer cell metastasis.28 Porcelli et al found that Notch pathway can accelerate cell cycle to enhance melanoma cell proliferation, which could antagonize the pharmacological anticancer effect of MEK inhibitor cobimetinib.26 Kaushik et al found that honokiol could inhibit melanomagenesis by suppressing Notch2 signaling.29 In our study, we found SOX10 could activate the transcription of Notch1, which can also upregulate the downstream protein NICD and Hes1 to activate the Notch signaling pathway (Figure 4D).
Although we found that SOX10 mRNA was highly expressed in melanoma, it was decreased in other tumor types (Figure 1A–F), such as BLCA, BRCA, CESC, COAD, ESCA, PRAD, READ and TGCT. Yin H indicated that SOX10 protein was overexpressed in BLCA, which could promote the BLCA progression.30 Moreover, Jamidi et al indicated that SOX10 protein has high level in BRCA, which could be a sensitive marker for BRCA patients.31 The main reason might lie in the existence of some post-transcriptional regulation for SOX10 in BLCA and BRCA. There are few studies on SOX10 in other tumors, so the function and mechanism of SOX10 are unclear in COAD, ESCA, PRAD, READ and TGCT. In our study, we found that SOX10 could activate the Notch pathway. Previous study indicated that Notch may act as an oncogene or a tumor suppressor, depending on the cellular environment.32 Therefore, the specific functional role of SOX10 in other tumors needs more basic studies to prove.
Conclusion
In summary, we found the transcriptional and post-transcriptional level of SOX10 were obviously increased in melanoma tissue samples, which negatively associated with patient’s prognosis. Moreover, SOX10 knockdown can inactive Notch signaling pathway to attenuate the proliferation ability of melanoma cell. This study indicated that inhibiting SOX10 and Notch pathway might be a potential therapeutic strategy in melanoma patients.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Wicklein D, Otto B, Suling A, et al. Ceacam1 promotes melanoma metastasis and is involved in the regulation of the emt associated gene network in melanoma cells. Sci Rep. 2018;8(1):11893. doi:10.1038/s41598-018-30338-4
2. Grzywa TM, Paskal W, Włodarski PK. Intratumor and intertumor heterogeneity in melanoma. Transl Oncol. 2017;10(6):956–975. doi:10.1016/j.tranon.2017.09.007
3. Harris ML, Baxter LL, Loftus SK, et al. Sox proteins in melanocyte development and melanoma. Pigment Cell Melanoma Res. 2010;23(4):496–513. doi:10.1111/j.1755-148X.2010.00711.x
4. Herbarth B, Pingault V, Bondurand N, et al. Mutation of the sry-related sox10 gene in dominant megacolon, a mouse model for human Hirschsprung disease. Proc Natl Acad Sci U S A. 1998;95(9):5161–5165. doi:10.1073/pnas.95.9.5161
5. Yu L, Peng F, Dong X, et al. Sex-determining region y chromosome-related high-mobility-group box 10 in cancer: a potential therapeutic target. Front Cell Dev Biol. 2020;8:564740. doi:10.3389/fcell.2020.564740
6. Murisier F, Guichard S, Beermann F. The tyrosinase enhancer is activated by sox10 and mitf in mouse melanocytes. Pigment Cell Res. 2007;20(3):173–184. doi:10.1111/j.1600-0749.2007.00368.x
7. Glatz K. [molecular heterogeneity of malignant melanomas]. Pathologe. 2007;28(6):474–478. German. doi:10.1007/s00292-007-0942-6
8. Shakhova O, Zingg D, Schaefer SM, et al. Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat Cell Biol. 2012;14(8):882–890. doi:10.1038/ncb2535
9. Cronin JC, Watkins-Chow DE, Incao A, et al. Sox10 ablation arrests cell cycle, induces senescence, and suppresses melanomagenesis. Cancer Res. 2013;73(18):5709–5718. doi:10.1158/0008-5472.CAN-12-4620
10. Graf SA, Busch C, Bosserhoff AK, et al. Sox10 promotes melanoma cell invasion by regulating melanoma inhibitory activity. J Invest Dermatol. 2014;134(8):2212–2220. doi:10.1038/jid.2014.128
11. Graf SA, Heppt MV, Wessely A, et al. The myelin protein pmp2 is regulated by sox10 and drives melanoma cell invasion. Pigment Cell Melanoma Res. 2019;32(3):424–434. doi:10.1111/pcmr.12760
12. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
13. Tomczak K, Czerwinska P, Wiznerowicz M. The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19(1A):A68–A77.
14. Tang Z, Li C, Kang B, et al. Gepia: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
15. Ormö M, Cubitt AB, Kallio K, et al. Crystal structure of the aequorea victoria green fluorescent protein. Science. 1996;273(5280):1392–1395. doi:10.1126/science.273.5280.1392
16. Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi:10.1126/science.1260419
17. Luo X, Xiong X, Shao Q, et al. The tumor suppressor interferon regulatory factor 8 inhibits β-catenin signaling in breast cancers, but is frequently silenced by promoter methylation. Oncotarget. 2017;8(30):48875–48888. doi:10.18632/oncotarget.16511
18. Uka R, Britschgi C, Krättli A, et al. Temporal activation of wnt/β-catenin signaling is sufficient to inhibit sox10 expression and block melanoma growth. Oncogene. 2020;39(20):4132–4154. doi:10.1038/s41388-020-1267-7
19. Willis BC, Johnson G, Wang J, et al. Sox10: a useful marker for identifying metastatic melanoma in sentinel lymph nodes. Appl Immunohistochem Mol Morphol. 2015;23(2):109–112. doi:10.1097/PAI.0000000000000097
20. Mohamed A, Gonzalez RS, Lawson D, et al. Sox10 expression in malignant melanoma, carcinoma, and normal tissues. Appl Immunohistochem Mol Morphol. 2013;21(6):506–510. doi:10.1097/PAI.0b013e318279bc0a
21. Ramos-Herberth FI, Karamchandani J, Kim J, et al. Sox10 immunostaining distinguishes desmoplastic melanoma from excision scar. J Cutan Pathol. 2010;37(9):944–952. doi:10.1111/j.1600-0560.2010.01568.x
22. Loftus SK. Decreased melanoma proliferation and cell survival: turn down your sox10. Pigment Cell Melanoma Res. 2013;26(1):3–4. doi:10.1111/pcmr.12028
23. Ayyildiz D, Piazza S. Introduction to bioinformatics. Methods Mol Biol. 2019;1986:1–15.
24. Ordóñez NG. Value of sox10 immunostaining in tumor diagnosis. Adv Anat Pathol. 2013;20(4):275–283. doi:10.1097/PAP.0b013e318297a9d0
25. Kontomanolis EN, Kalagasidou S, Pouliliou S, et al. The notch pathway in breast cancer progression. Scientific World J. 2018;2018:2415489. doi:10.1155/2018/2415489
26. Porcelli L, Mazzotta A, Garofoli M, et al. Active notch protects MAPK activated melanoma cell lines from mek inhibitor cobimetinib. Biomed Pharmacother. 2021;133:111006. doi:10.1016/j.biopha.2020.111006
27. Müller CS. Notch signaling and malignant melanoma. Adv Exp Med Biol. 2012;727:258–264.
28. Howard JD, Moriarty WF, Park J, et al. Notch signaling mediates melanoma-endothelial cell communication and melanoma cell migration. Pigment Cell Melanoma Res. 2013;26(5):697–707. doi:10.1111/pcmr.12131
29. Kaushik G, Venugopal A, Ramamoorthy P, et al. Honokiol inhibits melanoma stem cells by targeting notch signaling. Mol Carcinog. 2015;54(12):1710–1721. doi:10.1002/mc.22242
30. Yin H, Qin C, Zhao Y, et al. Sox10 is over-expressed in bladder cancer and contributes to the malignant bladder cancer cell behaviors. Clin Transl Oncol. 2017;19(8):1035–1044. doi:10.1007/s12094-017-1641-2
31. Jamidi SK, Hu J, Aphivatanasiri C, et al. Sry-related high-mobility-group/hmg box 10 (sox10) as a sensitive marker for triple-negative breast cancer. Histopathology. 2020;77(6):936–948. doi:10.1111/his.14118
32. Braune EB, Lendahl U. Notch – a goldilocks signaling pathway in disease and cancer therapy. Discov Med. 2016;21(115):189–196.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.