Back to Journals » International Journal of Nanomedicine » Volume 17
Soil Fungi as Biomediator in Silver Nanoparticles Formation and Antimicrobial Efficacy
Authors Sonbol H , Mohammed AE, Korany SM
Received 16 January 2022
Accepted for publication 12 April 2022
Published 29 June 2022 Volume 2022:17 Pages 2843—2863
DOI https://doi.org/10.2147/IJN.S356724
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Hana Sonbol,1 Afrah E Mohammed,1 Shereen M Korany2
1Department of Biology, College of Science, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia; 2Botany and Microbiology Department, Faculty of Science, Helwan University, Cairo, 11795, Egypt
Correspondence: Afrah E Mohammed, Department of Biology, College of Science, Princess Nourah bint Abdulrahman University, P.O. Box 84428, Riyadh, 11671, Saudi Arabia, Tel +966 566675853, Email [email protected]
Introduction and Objectives: Biogenic agents in nanoparticles fabrication are gaining great interest due to their lower possible negative environmental impacts. The present study aimed to isolate fungal strains from deserts in Saudi Arabia and assess their ability in silver nanoparticles (AgNPs) fabrication and evaluate their antibacterial effect.
Methods: Soil fungi were identified using 18s rDNA, and their ability in NPs fabrication was assessed as extracellular synthesis, then UV-vis spectroscopy, dynamic light scattering (DLS), energy-dispersive X-ray spectroscopy, and transmission electron microscopy were used for AgNPs characterization. The antibacterial activity of fungal-based NPs was assessed against one Gram-positive methicillin-resistant S. aureus (MRSA) and three Gram-negative bacteria (E. coli, Pseudomonas aeruginosa, and Klebsiella pneumoniae). Ultrastructural changes caused by fungal-based NPs on K. pneumoniae were investigated using TEM along with SDS-PAGE for protein profile patterns.
Results: The three fungal isolates were identified as Phoma sp. (MN995524), Chaetomium globosum (MN995493), and Chaetomium sp. (MN995550), and their filtrate reduced Ag ions into spherical P-AgNPs, G-AgNPs, and C-AgNPs, respectively. DLS data showed an average size between 12.26 and 70.24 nm, where EDX spectrums represent Ag at 3.0 keV peak. G-AgNPs displayed strong antibacterial activities against Klebsiella pneumoniae, and the ultrastructural changes caused by NPs were noted. Additionally, SDS-PAGE analysis of treated K. pneumoniae revealed fewer bands compared to control, which could be related to protein degradation.
Conclusion: Present findings have consequently developed an eco-friendly approach in NPs formation by environmentally isolated fungal strains to yield NPs as antibacterial agents.
Keywords: nanostructure, soil fungi, Phoma sp., Chaetomium sp., antibacterial, structural changes
Introduction
Recently growing bacterial resistance to antimicrobial drugs is a real danger to human health and has become one of the most serious matters in the medical sector. The antibiotics which are used mainly in the treatment of bacterial disease become defective over time or lead to the development of antibiotic-resistant strains.1 Consequently, it becomes a crucial demand to search for alternatives that help to overcome such challenges. Nanoparticles are used successfully in several medical and pharmaceutical applications,2 and such structures can be achieved by variable nanomaterials utilizing different metal ions like copper, magnesium, gold, and silver.3 Silver nanoparticles (AgNPs) can be spotlighted among the various forms of metallic nanoparticles for their wide antimicrobial potential.4,5 Because they are less reactive than silver ions, they may be better for clinical and therapeutic uses.6 In many fields, silver NPs are extensively used such as sensors, antimicrobial agents, filters, microelectronics, and catalysis in different areas, such as bio-labeling and efficiently against cancer cells as a cytotoxic agent due to their physicochemical and biological nature.7,8 Using silver nanoparticles for the control of different kinds of pathogenic microorganisms has been reported in many previous studies especially in the fields of health and agriculture.9 AgNPs have demonstrated effectiveness in killing both Gram-positive and Gram-negative bacteria10 with a multi-level mode of action since they have the ability to adhere to both cell walls and membranes of a bacterial cell passing into the interior cell structure and interact with DNA.11 They may also induce the formation of reactive oxygen species that damage biomacromolecules, and also alter the mechanisms of signal transduction.12,13 Conventional AgNPs synthesis approach may include chemical and physical agents that may lead to environmental toxicity and high energy consumption, and this has led to growing interest in biogenic synthesis approaches, as such an eco-friendly procedure enables obtaining nanoparticles with lower toxicity, better physicochemical characteristics, and greater stability.14 Biogenic formation of nanoparticles may be achieved by different organisms such as fungi, bacteria, and plants or even by its secondary metabolites which act as a reducing agent.15–17 Fungal extracts have been shown to yield numerous products for medical applications, for instance penicillin, statins, and cyclosporin, as well as mycotoxins, for example trichothecenes and aflatoxins, and with some anticancer activity.18,19 Therefore, fungi are considered as attractive agents for the biogenic synthesis of silver nanoparticles since they have excellent potential to produce many bioactive compounds, especially imperfect fungi and ascomycetes.20 This special capability of fungi may be attributed to their tolerance of heavy metals and their ability to bioaccumulate and internalize metals. Moreover, fungi can be cultivated easily on a large scale (“nanofactories”), producing high biomass, with the secretion of massive amounts of extracellular metabolites.21 Such produced biomolecules are not only used in the reduction process of metal ions but also help in capping the NPs providing better particle size and stability.22,23 A wide range of filamentous fungi and yeasts have also been recorded for their ability to synthesize silver nanoparticles, such as Phoma sp. 3.2883,24 Fusarium oxysporum,25 Aspergillus sp.,26 Beauveria bassiana,27 Penicillium sp.,28 Trichoderma harzianum,29 and Candida albicans.30 Also, Madbouly et al used Chaetomium globosum for the biosynthesis of AgNPs that showed in vivo antifungal activity against Fusarium wilt of tomato caused by Fusarium oxysporum f. sp. lycopersici.31 Each fungal species has its own ability in providing NPs with varied physicochemical and biological properties which might be related to their ability to produce a broad range of significant metabolites.32 Varied biomolecules from fungal extract were noted33 such as linoleic-acid derived psi factor from Aspergillus nidulans,33–36 zearalenone from Fusarium graminearum,37 butyrolactone I from Aspergillus terreus,38 melanin from Colletotrichum lagenarium, Alternaria alternate, and Cochliobolus heterotrophus,39–41 pigment from Aspergillus fumigatus,42 mycotoxins from Aspergillus spp.,43–45 and Patulin from Penicillium urticae.46 Furthermore, the endophytic C. globosum has been recorded from various niches as useful producers of bioactive agents. Several compounds have been isolated from endophytic C. globosum such as indole derivatives, chaetoglobosins,47 globosumones,48 globosuxanthone,49 cochliodes,50 azaphilones,51,52 and chaetoglocins.53 Several bioactive molecules were identified from Phoma sp. extracts such as phenolic compounds, triterpenes, steroids, reducing sugars.54 Furthermore, size of fungal-based NPs depends on the production conditions such as species, pH, temperature, and cultivation medium.55 The size of nanoparticles is considered one of the most important factors in determining their antimicrobial capability because smaller nanoparticles have greater effects.56 Antimicrobial activity of fungal-based NPs was noted earlier. Extracellular synthesis of Ag-NPs produced by Phoma glomerata (MTCC2210) has been evaluated and showed a broad bactericidal effect against Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa.57 The antimicrobial activity of endophytic C. globosum has been reported against different pathogenic bacteria and fungi, especially against plant pathogens.53,58,59 AgNPs synthesized by C. globosum JN711454 displayed an obvious antibacterial activity against many human pathogens such as Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli, and Klebsiella pneumoniae. Anti-inflammatory activity of AuNPs synthesized by Chaetomium globosum extract showed significant anti-inflammatory activity.60 Although varied studies used fungal extract for biosynthesis of NPs, information about soil-isolated and identified fungal strains as biogenic agents in NPs formation is lacking. Hereby, three soil fungal strains were obtained from different localities in Saudi Arabia and addressed for ability in AgNPs fabrication. Fungal isolates were identified based on 18S rDNA sequencing, and their accession numbers were deposited in GenBank. The main objectives of the work are to determine the potential of each fungus individually to synthesize AgNPs with a characterization of their physicochemical properties using UV-vis spectroscopy, FTIR, TEM, DLS, and EDX. Furthermore, NPs ability as antimicrobial agents was noted, and further ultrastructural changes and protein profiling of treated were assessed using TEM and SDS-PAGE consequently in a trial to detect the exact antibacterial mechanism of fungal-based NPs.
Materials and Methods
Isolation and Identification of Fungi
The three fungal isolates (Phoma sp., Chaetomium globosum, and Chaetomium sp.) used in this study were isolated from different sites of Saudi Arabia’s deserts. Fungi were isolated from soil at 5–20 cm depth using the dilution plate method.61 Samples were cultured on two different media, Potato Dextrose Agar (PDA), and Sabouraud Dextrose Agar with chloramphenicol 1%, and incubated for one week at 28 °C. Then, a serial inoculation plate was used to purify fungal isolates. Molecular identification using 18s rDNA was performed as described previously.62
Molecular Identification Using 18s rRNA Gene
In brief, fungal DNAs have been extracted using the InstaGeneTM Matrix Genomic DNA Kit (Bio-Rad Laboratories, Hercules, CA, USA) according to its manufacturer’s guidelines. A PCR assay of fungal 18s rRNA genes was performed using a DNA template and a couple of primers, NS1 F (5’ GTAGTCATATGCTTGTCTC 3’) and NS8 R (5’ TCCGCAGGTTCACCTACGGA 3’).63 The PCR reaction mixture consists of: 10X Taq PCR Buffer, 2 µL; 2.5 mM dNTP mixture, 1.6 µL; F and R primers (10 pmol/µL), 1.0 µL; KOMA Taq (2.5 U/µL), 0.2 µL; DNA template (20 ng/µL), 2 µL; and HPLC-grade distilled water to adjust the reaction volume to 20 µL. The amplification reactions were achieved in 20 µL using 1 µL of DNA template. The PCR was done as follows: initial denaturation was at 95 °C for 5 min, then 30 cycles including denaturation at 95 °C for 30s, annealing at 55 °C for 2 min, and extension at 68 °C for 1.5 min and a final extension at 68 °C for 10 minutes. The PCR products were confirmed using electrophoresis. Then, PCR purification was applied using the Montage PCR Cleanup Kit (MilliporeSigma, Burlington, MA, USA).
Sequencing and Analysis of the Amplified DNA
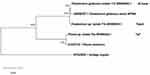
The purified PCR products were sequenced using the exact primers used for amplification with the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) and 3730xl DNA Analyzer automated DNA sequencing system (Applied Biosystems, USA) at Macrogen, Inc. (South Korea). The obtained 18s rRNA sequences of all fungal isolates were manually edited using Geneious prime software (Geneious Prime Version 2020.1.2).64 Consensus sequences were generated from forward and reverse sequences. The sequences obtained in this study were compared with their closely related reference strains in the National Center of Biotechnology Information (NCBI) database using the nucleotide BLASTn platform. A phylogenetic tree was constructed using the Neighbor-Joining method65 in MEGA X.66
Data Availability
The obtained nucleotide sequences of all three fungal isolates were deposited at GenBank with specific accession numbers (Table 1).
 |
Table 1 Isolated Fungi from Different Locations in Saudi Arabia |
Biomass and Filtrate Preparation
Two fungal discs of 4 mm from each isolate culture were inoculated into Sabouraud agar plates (Oxoid, UK) and incubated at 28 °C for seven days. Five discs (4 mm) were obtained from each plate, transferred to 500 mL Sabouraud broth (Oxoid, UK), and incubated at 28 °C for seven days. Biomass was collected through filtration using filter paper (Whatman no. 1), then washed with sterile distilled water for medium removal. Fungal preparations were weighed and added to distilled water, then incubated at 28 °C for 72 h. Finally, the water filtrates were obtained from the biomass31,67 and kept for further use.
Extracellular Synthesis of AgNPs
Prepared fungal filtrate was added to 1 mM of AgNO3 (Sigma Aldrich, UK) at a ratio of 1:1, then boiled for 30 min. The synthesis of AgNPs was performed under natural sunlight at 25 °C for 24 h. After the mixture color had changed to dark, the nanoparticles were separated by centrifugation at 14,000 rpm for 15 min. Thereafter, prepared NPs were taken for further characterization.
Ultraviolet-Visible Spectrophotometer Analysis
Ultraviolet-visible (UV-vis) spectroscopy absorption was determined using a spectrophotometer (© Shimadzu Corporation, UV-1800). The absorption spectra measurements for the NPs solution were performed within the range of 200–600 nm after 24 h of reaction using UV Probe software, and the fungal filtrate was used as a blank.31,67
Fourier Transform Infrared (FTIR) Spectroscopy
The FTIR spectrum detected the potential biomolecules in fungal filtrate responsible for reducing and capping the NPs. The measurement was recorded on FTIR spectroscopy (SPECTRUM100, Perkin-Elmer, USA) with a diffuse reflectance accessory, and the data scanning was done within the range 450–3500 cm−1.31,67
Dynamic Light Scattering (DLS)
The dynamic light scattering method used to assess the size distribution pattern was measured by a Zetasizer (NANO ZSP, Malvern Instruments Ltd, Serial Number: MAL1118778, ver 7.11, UK).31,67
Energy Dispersive X-Ray Spectroscopy (EDX)
EDX was used for the elemental analysis and confirmed the presence of the silver element accurately using SEM (JEOL, JED-2200 series, Japan).31,67
Transmission Electron Microscopy (TEM)
TEM was performed on a JEOL, Japan (JEM-1011) system to find the morphology and size distribution of AgNPs at 80 kV voltage. Samples were prepared by drop-coating on carbon-coated (200 mesh) TEM grids. The particle size distribution from TEM images was calculated using ImageJ software.
Antimicrobial Activity of Fungal-Based Silver Nanoparticles in vitro Assay
Four human pathogenic bacteria were used to examine the antibacterial activity of AgNPs including one Gram-positive methicillin-resistant S. aureus (MRSA) and three Gram-negative (E. coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa); they were obtained from the Bio-house medical lab, Riyadh, Saudi Arabia using the agar well diffusion method. Each strain was sub-cultured on nutrient agar medium (Oxoid) plates and grown for 24 h at 37 °C. Using a McFarland reader, a direct colony suspension method was applied, and McFarland standard (0.5) bacterial suspensions (1.5 × 108 CFU/mL) in the saline tube were prepared. The plates were inoculated by tested pathogenic strains. Twenty microliters of AgNPs were added separately into each well of the Petri plates and kept for drying under aseptic conditions. After 15 min, plates were incubated at 37 °C for 24 h. The inhibition zone surrounding the well was measured in mm. Antibacterial activity was determined, and zone diameter (mm) for antibiotics was recorded.68,69 The mycelial free extract was used to compare the antimicrobial activity of synthesized nanoparticles; also, ampicillin (1 mg/mL) was used as a positive control for bacterial strains. Then, the plates were incubated at 37 °C for 24 h, and the inhibition zone was determined in terms of a millimeter. These assays were performed in triplicate.
MICs and MBCs Determination
Minimal inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) were determined as per the method suggested by the National Committee of Laboratory Standards, using a 96-well plate. Four pathogenic bacteria strains, S. aureus, P. aeruginosa, Klebsiella pneumoniae, and E. coli, were used for detection of AgNPs antibacterial activity. Using the microdilution method in nutrient broth, approximately 2.5×105 CFU/mL (10 µL) of pathogenic bacteria was added individually to Nutrient broth (0.35 mL), and isolates were tested with different concentrations of AgNPs (0.031, 0.062, 0.125 and 0.250 mg/mL) and negative control composed of Nutrient broth inoculate with bacterial inoculum, and AgNPs solution composed of Nutrient broth with AgNPs were used. Then, 1 μL of samples from wells on Nutrient agar plates were subcultured and incubated for 24 h at 37 °C. MICs were evaluated by observing the difference between positive and negative control wells. MICs are known as the minimum concentration with low growth e.g. turbidity or pellet. Results are represented by the mean values of three independent replicates.68,69 On the other hand, to assess the minimum bactericidal concentrations (MBCs) the concentration that kills 99.9% of bacterial growth has been known as MBC.54,70,71 To measure the tolerance levels for the bacterial isolates used in this study with AgNPs concentration, MBCs/MICs ratios were calculated72 representative of the AgNPs' bactericidal capability against tested bacteria.73
SDS-PAGE for Protein Profile Pattern
Total cellular soluble proteins of K. pneumoniae extracted before and after treatment with 1 mg/mL of AgNPs for 24 h at 37 °C was purified by TriFast (Peqlab, VWR company) and further fractionated by OmniPAGE Mini vertical electrophoresis unit supplied with a power Pro 5 power supply (Cleaver Scientific, Warwickshire, UK) on a SERVAGel™ TG PRiME™ 10% (SERVA, Heidelberg, Germany).74
Bacterial Preparation for TEM
Pathogenic bacteria before and after treatment with G-AgNPs were cultivated in broth medium for 24 h at 37 °C, then centrifuged at 4000 rpm for 15 min, and the pellets resuspended in a mixture of 2.5% glutaraldehyde, 2% (para)formaldehyde in 100 mM cacodylate buffer (pH 7.0) with 2 mM CaCl2. After an initial ~30 min fixation, the specimens were cut into small (~1 mm3) pieces, followed by fixation in fresh fixative for 16–24 h at 4 °C. Specimens were washed briefly with 200 mM cacodylate buffer (pH 7.0) and post-fixed with 1% osmium tetroxide in 100 mM cacodylate buffer (pH 7.0); 2 h at 4 °C. They were then washed with excess distilled water for removal of any free cacodylate and/or phosphate ions and en bloc stained with 2.0% aqueous uranyl acetate, ~2 h at 4 °C (in dark). Dehydration was done with acetone (or ethanol) and propylene oxide, followed by embedding in resin as outlined above.75
Statistical Analysis
All tests were done in triplicate, and presented data are mean ± standard deviation (SD). One-way analysis of variance (ANOVA) and graph preparation were performed applying Prism 9.1 software (GraphPad Software Inc., La Jolla, CA, USA). Significance of the data is shown at p < 0.001 and p < 0.01. ImageJ (National Institutes of Health, Bethesda, MD, United States) was used for fungal-based NPs measurements.
Results
Isolation and Identification of Fungi
In the present study, three fungal isolates (Phoma sp., Chaetomium globosum, and Chaetomium sp.) were isolated from Saudi Arabia’s deserts and identified using molecular characterization by 18s rDNA sequencing.62 The 18s rRNA genes were amplified using universal primers, then sequence analysis was done by comparing the obtained sequence by NCBI-nBlast for constructing a phylogenetic tree (Figure 1, Table 1). Such isolates were used for the biosynthesis of AgNPs from AgNO3 and further described and examined for their antibacterial activities.
Biosynthesis of AgNPs
Filtrate from Phoma sp., Chaetomium globosum, and Chaetomium sp. was used for the AgNPs biosynthesis when added to AgNO3 and the mixtures kept at 25 °C under sunlight conditions. Color of the mixtures changed from colorless to stable brown color after 24 h for all tested fungal spp. (Figure 2), indicating NPs formation. Thereafter, the P-AgNPs, G-AgNPs, and C-AgNPs prepared from Phoma sp., Chaetomium globosum, and Chaetomium sp. filtrates, respectively, were taken for further analysis and description.
 |
Figure 2 Biosynthesis of AgNPs indicated by color change for the reaction medium composed of 1 mM AgNO3 and fungal filtrates after 24 h (left side) and fungal filtrate (right side). |
Characterization of Biogenic AgNPs
Reactions between Ag ions and fungal filtrate were noted by UV-vis spectroscopy that revealed absorbance at 450 nm, 400 nm, and 430 of the biosynthesized P-AgNPs, G-AgNPs, and C-AgNPs, respectively (Figure 3A–C).
 |
Figure 3 The UV-vis spectrum of the P-AgNPs (A), G-AgNPs (B), and C-AgNPs (C). |
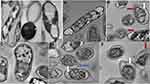
TEM imaging for P-AgNPs, G-AgNPs, and C-AgNPs demonstrated spherical NPs with no aggregation at average size of 12.7, 10.7, and 16.1 nm, respectively. Clear capping agents around AgNPs were also detected as light color (Figure 4). Smaller particle size was noted by TEM analysis using ImageJ software compared with that noted by DSL for the same NPs.
Analysis through EDX confirmed the presence of the silver element at 3 keV, along with a carbon peak and an oxygen peak (Figure 5). The Ag in P-AgNPs was 52.56%, in G-AgNPs was 28.87%, and that at C-AgNPs was 2.66%, and the peak clarity of EDX confirmed the purity of synthesized AgNPs.
 |
Figure 5 EDX of P-AgNPs (A), G-AgNPs (B), and C-AgNPs (C) indicating surface morphology quantitative analysis of silver atoms, carbon, and oxygen. |
Furthermore, P-AgNPs, G-AgNPs, and C-AgNPs showed average size diameters of 70.24, 62.13, and 12.26 nm, with a polydispersity index (PDI) of 0.298, 0.231, and 1.000, respectively (Figure 6). Furthermore, the particle size from intensity distribution showed major sizes of 98.41 nm (100%), 83.15 nm (96.5%), and 51.76 (100%) for P-AgNP, G-AgNPs, and C-AgNPs, respectively. Figure 6 Continued. Figure 6 The particle size distribution of P-AgNPs (A), G-AgNPs (B), and C-AgNPs (C).

FTIR spectroscopy was carried out to detect the organic molecules in the fungal filtrates and the AgNPs synthesized by their aids (Figure 7A–C). Absorbance peaks for the three fungal strains revealed two strong bands for each fungal filtrate. The signals of Phoma sp. filtrate were at 3259.51 and 1633.7 3cm−1 that moved to 3284.34 and 1635.4 cm−1 in P-AgNPs; and 3274.42 and 1634.25 cm−1 signals were noted for Chaetomium globosum filtrate that moved to 3294.44 and 1636.34 cm−1 in G-AgNPs. Furthermore, signals at 3260.43 and 1635.20 cm−1 were noted for Chaetomium sp. extract that moved to 3294.39 and 1638.52cm−1 in C-AgNPs. Different functional groups were noted in the fungal filtrate and the fungal-based NPs. Figure 7 Continued.
Antibacterial Activity of AgNPs
The antibacterial activities of fungal filtrate and fungal-based NPs were evaluated against four MDR pathogenic bacteria including S. aureus (MRSA) as well as P. aeruginosa, E. coli, and K. pneumoniae using the agar well diffusion method, and ampicillin was used as a positive control. No antibacterial activity was detected for all the fungal filtrate; however, fungal-based NPs inhibited the growth of both tested Gram-negative and Gram-positive strains (Table 2). The highest antibacterial activity of all fungal-based NPs was noticed against K. pneumoniae (Figure 8).
 |
Table 2 Inhibitory Effect (mm) of Fungal-Based NPs Against Some Strains of Pathogenic Bacteria |
The MIC and MBC were determined using the microdilution method. As shown in Table 3, the MICs (low turbidity or pellet formation in the well) were recorded at 0.031 and 0.062 mg/mL for G-AgNPs, for P-AgNPs the MIC was between 0.250 and 0.125 mg/mL, and for C-AgNPs the MIC was 0.125 mg/mL against all the bacterial strains tested. MBCs were observed at a concentration of 0.062 and 0.125 mg/mL for G-AgNPs, 0.25 and 0.5 mg/mL for P-AgNPs, and 0.25 mg/mL for C-AgNPs against all tested strains.
 |
Table 3 MICs and MBCs for the Biogenic AgNPs |
SDS-PAGE Banding Profile
In a trial to detect possible mechanism of NPs against K. pneumoniae, SDS-PAGE for protein profiling pattern before and after treatment with fungal-based NPs was performed (Figure 9) and indicated great variations. The pattern of protein detected for treated K. pneumoniae displayed lower protein band intensity in all treated bacteria in relation to untreated control. It was also noted that the number of detected bands was lower also in treated bacteria compared to control (Figure 9). Where lane (2) which shows untreated bacteria with a total of 14 bands ranging from 15 to 200 kDa, the number of bands in lane (3) shows 12 bands ranged from 16 to 122 kDa for K. pneumoniae treated with P-AgNPs. Furthermore, in lane (4) only 11 bands were noted from 17 to 134 kDa for K. pneumoniae treated with G-AgNPs. In the last lane (5) band number was reduced to 6 bands ranging from 26 to 140 kDa, which indicates a high level of polymorphism for K. pneumoniae treated with C-AgNPs. Such variations could be related to the protein degradation or loss and block in their synthesis pathways due to NPs treatment.
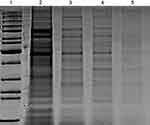 |
Figure 9 SDS-PAGE profile of protein extracted from K. pneumoniae (control) in lane (2) and treated bacteria with P-AgNPs, G-AgNPs, and C-AgNPs in lanes 3, 4, and 5 respectively. |
Ultrastructural Changes Detected by TEM
Furthermore, bacteria that showed high sensitivity to all prepared NPs was K. pneumoniae, therefore this strain was subjected G-AgNPs and noted under transmission electron microscope. Results of untreated K. pneumoniae showed that the cells kept their rod shape and the dense appearance of internal cytoplasmic material (Figure 10, Ctrl). However, treated K. pneumoniae exhibited different morphologies, including shrinking and thin cell wall, folded and light scattering cytoplasm, and reduction in cytoplasm. Generally, deformation of the treated cells was clearly observed (Figure 10).
Discussion
Multidrug resistance (MDR) bacteria is a topic of huge concern due to their growing worldwide incidences and the low efficacy of available drugs76,77 Bacteria exposed to an antibiotic might have various biological or structural alterations. The most important problems caused by antibiotic resistance mechanisms in pathogenic microorganisms are prevalence of bacterial mutations, horizontal gene transfer, destruction/alteration of antibiotic molecules, decrease in cell membrane permeability to prevent antibiotic penetration, higher expression of efflux pumps in the membrane, and alteration of antibiotic target sites.78 Therefore, environmentally sustainable, non-toxic, and cost-effective nano-therapeutics are in trend to treat such resistance mechanism in clinical bacteria. Few studies have reported the utilization of Phoma sp., Chaetomium globosum, and Chaetomium sp. for AgNPs production, even though the fungi population is a chemically rich organism possessing a wide range of compounds with promising anti-oxidant, anti-cancer, anti-inflammatory, and anti-microbial activities.79 Current study evaluated the three soil isolated fungi as biomediators in AgNPs formation by extracellular synthesis, and furthermore their efficiency as antibacterial agents was noted. Mechanism against K. pneumoniae was studied using TEM and SDS-PAGE analysis.
Biosynthesis of AgNPs
The biogenic materials in NPs formation provide capping agents (ie adsorption of molecules on the surface of NPs), which decides its final morphology and thus prevents the overgrowth of NPs.80,81 Generally, the biosynthesis of AgNPs follows the reduction of Ag+ ions to Ag0 from electrons liberated mainly from the biogenic agent’s active biomolecules that finally cap the formed NPs. Currently, the color of AgNO3 when mixed with each Phoma sp., Chaetomium globosum, and Chaetomium sp. filtrate changed drastically from colorless to dark brown, indicating the formation of AgNPs which could be well corroborated with earlier observations.24,31,57,60,82 Fungal filtrate interacted with Ag+ ions resulting in dark precipitation from the reaction mixture which could be due to surface plasmon resonance (SPR) owing to the collective oscillation of electrons.83 The possible mechanism for fungal filtrate in NPs formation could be related to the biomolecules that donate electrons to Ag+ ions which are therefore expected to form intermediate Ag-organic complexes, and the ions are then converted to Ag0 by free electrons produced in the medium.84 Repeated collisions among Ag0 atoms produce AgNPs that are meanwhile capped by other organics of fungal filtrate, giving specific size and shape to synthesized AgNPs. Furthermore, extracellular biogenic synthesis of nanoparticles is accomplished by fungal filtrate comprising only their bioactive molecules,85 such as citric acid, peroxidases, homogeneous proteins, and heterogeneous proteins and contributing to the stability of the formed nanoparticles.86 Fungal enzymes, especially nitrate reductases and electron shuttle quinones, or both, could be involved in the NPs production and stabilization mechanisms.87 However, further research and experimental trials are required to provide exact knowledge about the mechanism for the synthesis of nanoparticles using fungi.
Characterization of Biogenic AgNPs
Successful formation of AgNPs was further confirmed by UV-visible spectroscopy, DLS, EDX, and TEM analysis. AgNPs have optical features which give an initial idea about size, distribution, morphological shape, and surface properties, reducing agents, and stabilizers.88 The comparative UV-vis spectra identified showed a broad absorption at 400–450 nm of AgNPs with peaks overlapping the UV and visible regions for P-AgNPs, G-AgNPs, and C-AgNPs, which is the characteristic of AgNPs due to the transition of electrons.89 The absorption near to 400 nm by AgNPs has also been observed by other authors.57,90–92
The morphological analysis of fungal-based NPs through TEM at two different magnifications was performed. Aggregates of variable sizes NPs were recorded; however, the shape was monodispersed roughly spherical. Similar results obtained by Sathiyaraj et al92 revealed a variety of nanoparticle size obtained from Vallarai chooranam. Similar study by Elamawi et al93 showed variation of the obtained nanoparticle size from Trichoderma longibrachiatum which could be due to possible accumulation of proteins and enzymes that were secreted during the biosynthesis process.
The elemental composition of AgNPs revealed the presence of Ag with carbon and oxygen. The AgNPs after synthesis were washed many times to remove the ions and unbound fungal filtrate, therefore, the C and O appearing in EDX spectra could be from the fungal extract as described before.87 The peaks at 3.0 keV in the EDX spectrum represented Ag attributed to the SPR of Ag nanocrystals as reported earlier.92,94
Results obtained from DLS analysis revealed a wide base of recorded peaks referring to variable sizes of the formed AgNPs.93 The higher size detected for fungal-based NPs by DLS compared to the primary size measured by TEM displayed some sort of particle aggregation in the water solution which related to the frequency of NPs collisions as inter-particular interactions increase. As a result of these collisions, the average path length by NPs was reduced, thereby increasing the hydrodynamic size.95 Similar variation has been noted in another research where the size for PaNPs prepared by Delonix regia measured by DLS was 24.20 nm as compared to size 4 nm measured by TEM.96 A similar kind of variation has been also noted in another study where size of hydrodynamic diameter of AgNPs prepared by T. asperellum was 1.2-fold higher than that measured by TEM.97
The FTIR spectrum of AgNPs suggests the chemical structure of NPs through the identification of functional groups attached to their surface with absorption pattern differing from free ones.87 In this study, the signals of FTIR detected for fungal filtrate and NPs are assigned to different functional groups. All the major signals identified for fungal filtrate were also noted in fungal-based NPs; however, slight alterations in the position and the strength of bands of absorption were noted and indicated contribution of fungal bioactive molecules in the formation of NPs. The signals for all fungal filtrate and fungal-based NPs suggest the involvement of aliphatic hydrocarbons, aromatic rings, and aliphatic amines in the process of bio-reduction.98,99 The two peaks that appeared in the diffraction gram may be due to the presence of reducing and capping organic moieties attached to AgNPs as reported by other researchers.100,101 The bands of FTIR from 1633.79 to 1638.52 cm−1 may correspond to the C=C aromatic ring stretch.99 In addition, the bands of FTIR from 3060.43 to 3294.39 cm−1, could be attributed to the presence of polyphenolic –OH groups.102 The probable role of OH functional group-containing molecules such as polyols consisting of terpenoids, tannins, saponins, etc.102 is forming a complex with fungal-based NPs as shown by reduction in the peaks. In addition, the observed shift in transmittance of FTIR peaks of fungal-based NPs compared to fungal filtrates could be due to probable interactions of functional groups from the filtrate with metal ions (in reduction process) and NPs (through capping).103
Antibacterial Activity of AgNPs
The antibacterial activity of fungal filtrate and fungal-based NPs was evaluated against four pathogenic bacteria. No antibacterial activity was detected for all fungal filtrates; however, K. pneumoniae was the most affected human pathogen by G-AgNPs, and the lowest antibacterial activity of all fungal-based NPs was noticed against E.coli and P. aeruginosa, and the highest antibacterial effect detected for P-AgNPs was against tested E.coli bacteria. Variations in the zone of inhibition can be explained by the structure of the Gram-positive and Gram-negative bacteria cell wall. The cell wall in Gram-positive bacteria consists of a thick peptidoglycan coating, which is made up of short peptide cross-linked linear polysaccharide chains that cause rigid construction of the cell wall, whereas the cell wall of Gram-negative bacteria contain a thin peptidoglycan layer that is more easily damaged.92,104
Antibacterial activity of AgNPs synthesized using Plumeria pudica Jacq. flower extract showed the highest inhibition against Gram-negative bacteria E. coli.90 Antibacterial activity of the AgNPs synthesized using Jatropha integerrima Jacq. flower extract exhibits high growth inhibitory effect towards E. coli which was low against B. subtilis.91 Along with our results, a study by Durán et al105 reported a reduction of Ag cations by C. globosum with subsequent high antimicrobial activity. Furthermore, AgNPs prepared by C. globosum showed antifungal activity against pathogenic F. oxysporum.31 A similar kind of bacteria was used by Sathiyaraj et al92 to examine the antibacterial characteristic of Vallarai chooranam synthesized AgNPs.
AgNPs are supposed to exhibit antibacterial activity by the formation of reactive oxygen species which interact with glycoprotein on the bacterial cell wall then get onto the cytoplasm.106 The noted activity of all fungal-based NPs against K. pneumoniae might be due to the structure of cell wall of Gram-negative bacteria.107
Regarding MIC and MBC, a recent study by Lotfy et al94 reported a similar kind of observations when they studied NPs mediated by Aspergillus terreus against twelve bacteria, where P. aeruginosa and S. faecalis showed the highest susceptibility. Similar research by Sathiyaraj et al92 revealed that AgNPs prepared by Vallarai chooranam showed a high zone of inhibition against E. coli and low one against Bacillus subtilis. Current results regarding MBC:MIC indicated that tested strains were non tolerant. 72 Variations in the MIC and MBC values of NPs in this work could be due to particle size, the nature of the material coating the NPs, and the response of treated bacteria.108,109
SDS-PAGE Banding Profile
SDS-PAGE for protein profiling pattern of K. pneumoniae before and after treatment with fungal-based NPs showed great variations, which indicates a high level of polymorphism. The SDS-PAGE pattern of bacteria treated with NPs showed low band intensity of the proteins. NPs might cause extreme oxidative stress, producing unfolding of the protein chain, which leads to protein changes and degradation.110,111 In addition, presence of new protein bands detected after K. pneumoniae exposed to C-AgNPs could be related to the development of new proteins as response to stress.112,113
K. pneumoniae Ultrastructural Changes
TEM micrographs of K. pneumoniae showed the presence of membrane blebbing, separation of cytoplasmic membrane from cell wall, and rupture in the outer membrane after being subjected to G-AgNPs. Similar results were noted by Veras et al,114 who showed different morphological and ultrastructural changes on the K. pneumoniae isolates after these were exposed to β-lactams from diverse concentrations, for example cell filamentation, loss of cytoplasmic material, and disorganization of the wall. In addition, the treated bacteria became elongated, and reduction in the cytoplasmic material and existence of membrane components were observed suggesting that bacteria have been affected by G-AgNPs in a similar pattern to low concentrations of some antibiotics.115 Similar findings were noted by DeLoney and Schiller115 which showed that Helicobacter pylori was turned to a filamentous shape after being exposed to aztreonam and to a spherical shape when exposed to other β-lactams. Furthermore, electron-lucent space with different sizes and shapes in the cytoplasm was observed, and some NPs that appeared in the cell wall and inside the cell were noted earlier.114 Understanding the effect of G-AgNPs on K. pneumoniae isolates is significant because this permits understanding of how the therapeutic mixtures can efficiently help towards treating the infections initiated by these isolates. Generally, variations in antimicrobial activity of fungal-based NPs prepared here could not be highly significant which might be due to comparable characteristics for each NP, leading to similar biological behavior. K. pneumoniae was sensitive to all fungal-based NPs, which could be related to their cell wall structure; however, not all tested Gram-negative bacteria followed the same pattern.
Conclusion
The present research has been developed in an eco-friendly approach in NPs formation by using soil-isolated fungal strains Phoma sp., Chaetomium globosum, and Chaetomium sp. with antibacterial activity against four human pathogenic bacterial strains. Spherical NPs were noted by TEM, and hydrodynamic radius of 70.24, 62.13, and 12.26 nm average diameters for P-AgNPs, G-AgNPs, and C-AgNPs, respectively, were noted. K. pneumoniae was the most affected bacterial strain with fungal-based NPs and was studied by TEM analysis before and after treatment showing morphological changes indicating deformation of cellular structure via direct interaction between NPs and cellular components. Also, SDS-PAGE for protein profiling pattern of K. pneumoniae before and after treatment with fungal-based NPs showed great variations, which indicates a high level of polymorphism. In addition, presence of new protein bands detected after K. pneumoniae was exposed to NPs could be attributed to formation of stress response protein. However, further investigations are required to validate this presumption by screening fungal-based NPs biological activities. The current study gives a good picture on the variations in antimicrobial activity of fungal-based NPs.
Data Sharing Statement
Data of this manuscript are displayed in Figures 1–10. The facts and raw data analyzed are available from the corresponding author upon request.
Acknowledgments
This research has been supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R83), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Ipe DS, Kumar PT, Love RM, Hamlet SM. Silver nanoparticles at biocompatible dosage synergistically increases bacterial susceptibility to antibiotics. Front Microbiol. 2020;11:1074. doi:10.3389/fmicb.2020.01074
2. Makarov VV, Love AJ, Sinitsyna OV, et al. “Green” nanotechnologies: synthesis of metal nanoparticles using plants. Acta Naturae. 2014;6(1):20. doi:10.32607/20758251-2014-6-1-35-44
3. Hemath Naveen KS, Kumar G, Karthik L, Bhaskara Rao KV. Extracellular biosynthesis of silver nanoparticles using the filamentous fungus Penicillium sp. Arch Appl Sci Res. 2010;2(6):161–167.
4. Loo YY, Rukayadi Y, Nor-Khaizura MAR, et al. In vitro antimicrobial activity of green synthesized silver nanoparticles against selected gram-negative foodborne pathogens. Front Microbiol. 2018;9:1555. doi:10.3389/fmicb.2018.01555
5. Aabed K, Mohammed AE. Synergistic and antagonistic effects of biogenic silver nanoparticles in combination with antibiotics against some pathogenic microbes. Front Bioeng Biotechnol. 2021;9. doi:10.3389/fbioe.2021.652362
6. Madhumathi K, Kumar PTS, Abhilash S, et al. Development of novel chitin/nanosilver composite scaffolds for wound dressing applications. J Mater Sci Mater Med. 2010;21(2):807–813. doi:10.1007/s10856-009-3877-z
7. Kim YS, Kim JS, Cho HS, et al. Twenty-eight-day oral toxicity, genotoxicity, and gender-related tissue distribution of silver nanoparticles in Sprague-Dawley rats. Inhal Toxicol. 2008;20(6):575–583. doi:10.1080/08958370701874663
8. Rai M, Yadav A, Gade A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv. 2009;27(1):76–83. doi:10.1016/j.biotechadv.2008.09.002
9. Burdușel A-C, Gherasim O, Grumezescu AM, Mogoantă L, Ficai A, Andronescu E. Biomedical applications of silver nanoparticles: an up-to-date overview. Nanomaterials. 2018;8(9):681. doi:10.3390/nano8090681
10. Taglietti A, Diaz Fernandez YA, Amato E, et al. Antibacterial activity of glutathione-coated silver nanoparticles against gram positive and gram negative bacteria. Langmuir. 2012;28(21):8140–8148. doi:10.1021/la3003838
11. Li W-R, Xie X-B, Shi Q-S, Duan -S-S, Ouyang Y-S, Chen Y-B. Antibacterial effect of silver nanoparticles on Staphylococcus aureus. Biometals. 2011;24(1):135–141. doi:10.1007/s10534-010-9381-6
12. Dakal TC, Kumar A, Majumdar RS, Yadav V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front Microbiol. 2016;7:1831. doi:10.3389/fmicb.2016.01831
13. Kim SW, Jung JH, Lamsal K, Kim YS, Min JS, Lee YS. Antifungal effects of silver nanoparticles (AgNPs) against various plant pathogenic fungi. Mycobiology. 2012;40(1):53–58. doi:10.5941/MYCO.2012.40.1.053
14. Iravani S, Korbekandi H, Mirmohammadi SV, Zolfaghari B. Synthesis of silver nanoparticles: chemical, physical and biological methods. Res Pharm Sci. 2014;9(6):385.
15. Durán N, Marcato PD, Durán M, Yadav A, Gade A, Rai M. Mechanistic aspects in the biogenic synthesis of extracellular metal nanoparticles by peptides, bacteria, fungi, and plants. Appl Microbiol Biotechnol. 2011;90(5):1609–1624. doi:10.1007/s00253-011-3249-8
16. Mohammed AE, Al-Keridis LA, Rahman I, et al. Silver nanoparticles formation by jatropha integerrima and LC/MS-QTOF-based metabolite profiling. Nanomaterials. 2021;11(9):2400. doi:10.3390/NANO11092400
17. Kumar LH, Kazi SN, Masjuki HH, Zubir MNM. A review of recent advances in green nanofluids and their application in thermal systems. Chem Eng J. 2022;429:132321. doi:10.1016/j.cej.2021.132321
18. Chandra S. Endophytic fungi: novel sources of anticancer lead molecules. Appl Microbiol Biotechnol. 2012;95(1):47–59. doi:10.1007/s00253-012-4128-7
19. Keller NP, Turner G, Bennett JW. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol. 2005;3(12):937–947. doi:10.1038/nrmicro1286
20. Berdy J. Bioactive microbial metabolites. J Antibiot. 2005;58(1):1–26. doi:10.1038/ja.2005.1
21. Li Q, Liu F, Li M, Chen C, Gadd GM. Nanoparticle and nanomineral production by fungi. Fungal Biol Rev. 2021. doi:10.1016/j.fbr.2021.07.003
22. Azmath P, Baker S, Rakshith D, Satish S. Mycosynthesis of silver nanoparticles bearing antibacterial activity. Saudi Pharm J. 2016;24(2):140–146. doi:10.1016/j.jsps.2015.01.008
23. Adebayo EA, Azeez MA, Alao MB, Oke AM, Aina DA. Fungi as veritable tool in current advances in nanobiotechnology. Heliyon. 2021;7(11):e08480. doi:10.1016/j.heliyon.2021.e08480
24. Chen JC, Lin ZH, Ma XX. Evidence of the production of silver nanoparticles via pretreatment of Phoma sp. 3.2883 with silver nitrate. Lett Appl Microbiol. 2003;37(2):105–108. doi:10.1046/j.1472-765X.2003.01348.x
25. Vijayan S, Divya K, George TK, Jisha MS. Biogenic synthesis of silver nanoparticles using endophytic fungi Fusarium oxysporum isolated from Withania somnifera (L.), its antibacterial and cytotoxic activity. J Bionanosci. 2016;10(5):369–376. doi:10.1166/jbns.2016.1390
26. Zomorodian K, Pourshahid S, Sadatsharifi A, et al. Biosynthesis and characterization of silver nanoparticles by aspergillus species. Biomed Res Int. 2016;2016:1–6. doi:10.1155/2016/5435397
27. Kamil D, Prameeladevi T, Ganesh S, Prabhakaran N, Nareshkumar R, Thomas SP. Green synthesis of silver nanoparticles by entomopathogenic fungus Beauveria bassiana and their bioefficacy against mustard aphid (Lipaphis erysimi Kalt.). Current Drug Delivery. 2017;14:816–831. doi:10.2174/1567201813666160919142212
28. Singh D, Rathod V, Ninganagouda S, Hiremath J, Singh AK, Mathew J. Optimization and characterization of silver nanoparticle by endophytic fungi Penicillium sp. isolated from Curcuma longa (Turmeric) and application studies against MDR E. coli and S. aureus. Bioinorg Chem Appl. 2014;2014:1–8. doi:10.1155/2014/408021
29. Guilger M, Pasquoto-Stigliani T, Bilesky-Jose N, et al. Biogenic silver nanoparticles based on Trichoderma harzianum: synthesis, characterization, toxicity evaluation and biological activity. Sci Rep. 2017;7(1):1–13. doi:10.1038/srep44421
30. Bhat MA, Nayak BK, Nanda A. Evaluation of bactericidal activity of biologically synthesised silver nanoparticles from Candida albicans in combination with ciprofloxacin. Mater Today Proc. 2015;2(9):4395–4401. doi:10.1016/j.matpr.2015.10.036
31. Madbouly AK, Abdel-Aziz MS, Abdel-Wahhab MA. Biosynthesis of nanosilver using Chaetomium globosum and its application to control Fusarium wilt of tomato in the greenhouse. Iet Nanobiotechnol. 2017;11(6):702–708. doi:10.1049/iet-nbt.2016.0213
32. Dhillon GS, Brar SK, Kaur S, Verma M. Green approach for nanoparticle biosynthesis by fungi: current trends and applications. Crit Rev Biotechnol. 2012;32(1):49–73. doi:10.3109/07388551.2010.550568
33. Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev. 2002;66(3):447–459. doi:10.1128/MMBR.66.3.447-459.2002
34. Champe SP, El-Zayat AA. Isolation of a sexual sporulation hormone from Aspergillus nidulans. J Bacteriol. 1989;171(7):3982–3988. doi:10.1128/jb.171.7.3982-3988.1989
35. Champe SP, Rao P, Chang A. An endogenous inducer of sexual development in Aspergillus nidulans. Microbiology. 1987;133(5):1383–1387. doi:10.1099/00221287-133-5-1383
36. EbrahimáEl-Zayat AA. Structure and synthesis of sporogenic psi factors from Aspergillus nidulans. J Chem Soc Chem Commun. 1991;(20):1486–1487. doi:10.1039/C39910001486
37. Wolf JC, Mirocha CJ. Regulation of sexual reproduction in Gibberella zeae (Fusarium roseum’Graminearum’) by F-2 (zearalenone). Can J Microbiol. 1973;19(6):725–734. doi:10.1139/m73-117
38. Schimmel TG, Coffman AD, Parsons SJ. Effect of butyrolactone I on the producing fungus, Aspergillus terreus. Appl Environ Microbiol. 1998;64(10):3707–3712. doi:10.1128/AEM.64.10.3707-3712.1998
39. Takano Y, Kikuchi T, Kubo Y, Hamer JE, Mise K, Furusawa I. The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol Plant Microbe Interact. 2000;13(4):374–383. doi:10.1094/MPMI.2000.13.4.374
40. Kawamura C, Tsujimoto T, Tsuge T. Targeted disruption of a melanin biosynthesis gene affects conidial development and UV tolerance in the Japanese pear pathotype of Alternaria alternata. Mol Plant Microbe Interact. 1999;12(1):59–63. doi:10.1094/MPMI.1999.12.1.59
41. Leonard KJ. Virulence, temperature optima, and competitive abilities of isolines of races T and O of Bipolaris maydis. Phytopathology. 1977;67(11):1273–1279. doi:10.1094/Phyto-67-1273
42. Tsai HF, Chang YC, Washburn RG, Wheeler MH, Kwon-Chung KJ. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J Bacteriol. 1998;180(12):3031–3038. doi:10.1128/jb.180.12.3031-3038.1998
43. Bennett JW, Papa KE. The aflatoxigenic Aspergillus spp. Adv Plant Pathol Genet Plant Pathog Fungi. 1988;6:263–280.
44. Guzmán-de-peña D, Aguirre J, Ruiz-Herrera J. Correlation between the regulation of sterigmatocystin biosynthesis and asexual and sexual sporulation in Emericella nidulans. Antonie Van Leeuwenhoek. 1998;73(2):199–205. doi:10.1023/A:1000820221945
45. Reiß J. Development of Aspergillus parasiticus and formation of aflatoxin B 1 under the influence of conidiogenesis affecting compounds. Arch Microbiol. 1982;133(3):236–238. doi:10.1007/BF00415008
46. Sekiguchi J, Gaucher GM. Conidiogenesis and secondary metabolism in Penicillium urticae. Appl Environ Microbiol. 1977;33(1):147–158. doi:10.1128/aem.33.1.147-158.1977
47. Jiao W, Feng Y, Blunt JW, Cole ALJ, Munro MHG, Chaetoglobosins Q. R, and T, three further new metabolites from Chaetomium g lobosum. J Nat Prod. 2004;67(10):1722–1725. doi:10.1021/np030460g
48. Bashyal BP, Wijeratne EMK, Faeth SH, Gunatilaka AAL. Globosumones A− C, cytotoxic orsellinic acid esters from the Sonoran desert endophytic fungus Chaetomium globosum. J Nat Prod. 2005;68(5):724–728. doi:10.1021/np058014b
49. Wijeratne EMK, Turbyville TJ, Fritz A, Whitesell L, Gunatilaka AAL. A new dihydroxanthenone from a plant-associated strain of the fungus Chaetomium globosum demonstrates anticancer activity. Bioorg Med Chem. 2006;14(23):7917–7923. doi:10.1016/j.bmc.2006.07.048
50. Debbab A, Aly AH, Edrada-Ebel R, et al. Bioactive secondary metabolites from the endophytic fungus Chaetomium sp. isolated from Salvia officinalis growing in Morocco. Biotechnol Agron Soc Environ. 2009;13(2):229–234.
51. Yasuhide M, Yamada T, Numata A, Tanaka R. Chaetomugilins, new selectively cytotoxic metabolites, produced by a marine fish-derived Chaetomium species. J Antibiot. 2008;61(10):615–622. doi:10.1038/ja.2008.81
52. Borges WS, Mancilla G, Guimaraes DO, Durán-Patrón R, Collado IG, Pupo MT. Azaphilones from the endophyte Chaetomium globosum. J Nat Prod. 2011;74(5):1182–1187. doi:10.1021/np200110f
53. Ge HM, Zhang Q, Xu SH, et al. Chaetoglocins A–D, four new metabolites from the endophytic fungus Chaetomium globosum. Planta Med. 2011;77(03):277–280. doi:10.1055/s-0030-1250292
54. da Silva GB, Silvino KF, Bezerra JDP, De farias TGS, de Araã°jo JM, Stamford TLM. Antimicrobial activity of Phoma sp. URM 7221: an endophyte from Schinus terebinthifolius Raddi (Anacardiaceae). African J Microbiol Res. 2017;11(1):1–7. doi:10.5897/AJMR2016.8326
55. Lee SH, Jun B-H. Silver nanoparticles: synthesis and application for nanomedicine. Int J Mol Sci. 2019;20(4):865. doi:10.3390/ijms20040865
56. Lu Z, Rong K, Li J, Yang H, Chen R. Size-dependent antibacterial activities of silver nanoparticles against oral anaerobic pathogenic bacteria. J Mater Sci Mater Med. 2013;24(6):1465–1471. doi:10.1007/s10856-013-4894-5
57. Birla SS, Tiwari VV, Gade AK, Ingle AP, Yadav AP, Rai MK. Fabrication of silver nanoparticles by Phoma glomerata and its combined effect against Escherichia coli, Pseudomonas aeruginosa and Staphylococcus aureus. Lett Appl Microbiol. 2009;48(2):173–179. doi:10.1111/j.1472-765X.2008.02510.x
58. Marwah RG, Fatope MO, Deadman ML, Al-Maqbali YM, Husband J. Musanahol: a new aureonitol-related metabolite from a Chaetomium sp. Tetrahedron. 2007;63(34):8174–8180. doi:10.1016/j.tet.2007.05.119
59. Park J-H, Choi GJ, Jang KS, et al. Antifungal activity against plant pathogenic fungi of chaetoviridins isolated from Chaetomium globosum. FEMS Microbiol Lett. 2005;252(2):309–313. doi:10.1016/j.femsle.2005.09.013
60. Ningaraju S, Munawer U, Raghavendra VB, et al. Chaetomium globosum extract mediated gold nanoparticle synthesis and potent anti-inflammatory activity. Anal Biochem. 2021;612:113970. doi:10.1016/j.ab.2020.113970
61. Waksman SA. A method for counting the number of fungi in the soil. J Bacteriol. 1922;7(3):339. doi:10.1128/jb.7.3.339-341.1922
62. Alotaibi MO, Sonbol HS, Alwakeel SS, et al. Microbial diversity of some sabkha and desert sites in Saudi Arabia. Saudi J Biol Sci. 2020;27:2778–2789. doi:10.1016/j.sjbs.2020.06.038
63. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc Guid Methods Appl. 1990;18(1):315–322.
64. Kearse M, Moir R, Wilson A, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi:10.1093/bioinformatics/bts199
65. Saitou N, Masatoshi N. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi:10.1093/oxfordjournals.molbev.a040454
66. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi:10.1093/molbev/msy096
67. Rai M, Ingle AP, Gade AK, Duarte MCT, Duran N. Three Phoma spp. synthesised novel silver nanoparticles that possess excellent antimicrobial efficacy. IET Nanobiotechnol. 2015;9(5):280–287. doi:10.1049/iet-nbt.2014.0068
68. CLSI. Performance Standards for Antimicrobial Susceptibility Testing CLSI Supplement M100S. Wayne, PA: Clin Lab Stand Institute; 2016.
69. CLSI. M02-A11: performance standards for antimicrobial disk susceptibility tests; approved standard. Clin Lab Stand Inst. 2015;32(1):92.
70. Das P, Xenopoulos MA, Williams CJ, Hoque ME, Metcalfe CD. Effects of silver nanoparticles on bacterial activity in natural waters. Environ Toxicol Chem. 2012;31(1):122–130. doi:10.1002/etc.716
71. Basri DF, Fan SH. The potential of aqueous and acetone extracts of galls of Quercus infectoria as antibacterial agents. Indian J Pharmacol. 2005;37(1):26. doi:10.4103/0253-7613.13851
72. May J, Shannon K, King A, French G. Glycopeptide tolerance in Staphylococcus aureus. J Antimicrob Chemother. 1998;42(2):189–197. doi:10.1093/jac/42.2.189
73. Mohammed AE, Al-Qahtani A, Al-Mutairi A, Al-Shamri B, Aabed K. Antibacterial and cytotoxic potential of biosynthesized silver nanoparticles by some plant extracts. Nanomaterials. 2018;8(6):382. doi:10.3390/nano8060382
74. Hamida RS, Abdelmeguid NE, Ali MA, Bin-Meferij MM, Khalil MI. Synthesis of silver nanoparticles using a novel cyanobacteria Desertifilum sp. extract: their antibacterial and cytotoxicity effects. Int J Nanomedicine. 2020;15:49. doi:10.2147/IJN.S238575
75. Glauert AM, Lewis PR. Biological Specimen Preparation for Transmission Electron Microscopy.
76. Klemm EJ, Wong VK, Dougan G. Emergence of dominant multidrug-resistant bacterial clades: lessons from history and whole-genome sequencing. Proc Natl Acad Sci USA. 2018;115:12872–12877. doi:10.1073/pnas.1717162115
77. Nikolaou M, Pavlopoulou A, Georgakilas AG, Kyrodimos E. The challenge of drug resistance in cancer treatment: a current overview. Clin Exp Metastasis. 2018;35:309–318. doi:10.1007/s10585-018-9903-0
78. Munita JM, Arias CA, Unit AR, De Santiago A. HHS public access mechanisms of antibiotic resistance. HHS Public Access. 2016;4(2):1–37. doi:10.1128/microbiolspec.VMBF-0016-2015.Mechanisms
79. Gaikwad S, Ingle A, Gade A, et al. Antiviral activity of mycosynthesized silver nanoparticles against herpes simplex virus and human parainfluenza virus type 3. Int J Nanomedicine. 2013;8:4303. doi:10.2147/IJN.S50070
80. Sonbol H, Ameen F, AlYahya S, Almansob A, Alwakeel S. Padina boryana mediated green synthesis of crystalline palladium nanoparticles as potential nanodrug against multidrug resistant bacteria and cancer cells. Sci Rep. 2021;11(1):5444. doi:10.1038/s41598-021-84794-6
81. Vaseghi Z, Nematollahzadeh A, Tavakoli O. Green methods for the synthesis of metal nanoparticles using biogenic reducing agents: a review. Rev Chem Eng. 2018;34(4):529–559. doi:10.1515/revce-2017-0005
82. Siddiqi KS, Husen A, Rao RAK. A review on biosynthesis of silver nanoparticles and their biocidal properties. J Nanobiotechnol. 2018;16(1):1–28. doi:10.1186/s12951-017-0328-8
83. Singh A, Jain D, Upadhyay MK, Khandelwal N, Verma HN. Green synthesis of silver nanoparticles using Argemone mexicana leaf extract and evaluation of their antimicrobial activities. Dig J Nanomater Bios. 2010;5(2):483–489.
84. Basavaraja S, Balaji SD, Lagashetty A, Rajasab AH, Venkataraman A. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium semitectum. Mater Res Bull. 2008;43(5):1164–1170. doi:10.1016/j.materresbull.2007.06.020
85. Khan NT, Khan MJ, Jameel J, Jameel N, Rheman SUA. An overview: biological organisms that serves as nanofactories for metallic nanoparticles synthesis and fungi being the most appropriate. Bioceram Dev Appl. 2017;7:101. doi:10.4172/2090-5025.1000101
86. Netala VR, Bethu MS, Pushpalatha B, et al. Biogenesis of silver nanoparticles using endophytic fungus Pestalotiopsis microspora and evaluation of their antioxidant and anticancer activities. Int J Nanomedicine. 2016;11:5683. doi:10.2147/IJN.S112857
87. Khandel P, Shahi SK. Mycogenic nanoparticles and their bio-prospective applications: current status and future challenges. J Nanostruct Chem. 2018;8(4):369–391. doi:10.1007/s40097-018-0285-2
88. Abou El-Nour KMM, Eftaiha A, Al-Warthan A, Ammar RAA. Synthesis and applications of silver nanoparticles. Arab J Chem. 2010;3(3):135–140. doi:10.1016/j.arabjc.2010.04.008
89. Mie R, Samsudin MW, Din LB, Ahmad A, Ibrahim N, Adnan SNA. Synthesis of silver nanoparticles with antibacterial activity using the lichen Parmotrema praesorediosum. Int J Nanomedicine. 2014;9:121. doi:10.2147/IJN.S52306
90. Suriyakala G, Sathiyaraj S, Gandhi AD, Vadakkan K, Mahadeva Rao US, Babujanarthanam R. Plumeria pudica Jacq. flower extract - mediated silver nanoparticles: characterization and evaluation of biomedical applications. Inorg Chem Commun. 2021;126:108470. doi:10.1016/j.inoche.2021.108470
91. Suriyakala G, Sathiyaraj S, Devanesan S, et al. Phytosynthesis of silver nanoparticles from Jatropha integerrima Jacq. flower extract and their possible applications as antibacterial and antioxidant agent. Saudi J Biol Sci. 2022;29(2):680–688. doi:10.1016/j.sjbs.2021.12.007
92. Sathiyaraj S, Suriyakala G, Gandhi AD, et al. Green biosynthesis of silver nanoparticles using vallarai chooranam and their potential biomedical applications. J Inorg Organomet Polym Mater. 2020;30(11):4709–4719. doi:10.1007/s10904-020-01683-7
93. Elamawi RM, Al-Harbi RE, Hendi AA. Biosynthesis and characterization of silver nanoparticles using Trichoderma longibrachiatum and their effect on phytopathogenic fungi. Egypt J Biol Pest Control. 2018;28(1):1–11. doi:10.1186/s41938-018-0028-1
94. Lotfy WA, Alkersh BM, Sabry SA, Ghozlan HA. Biosynthesis of silver nanoparticles by Aspergillus terreus: characterization, optimization, and biological activities. Front Bioeng Biotechnol. 2021;9. doi:10.3389/fbioe.2021.633468
95. Bhattacharjee S. DLS and zeta potential - what they are and what they are not? J Control Release. 2016;235:337–351. doi:10.1016/j.jconrel.2016.06.017
96. Dauthal P, Mukhopadhyay M. Biosynthesis of palladium nanoparticles using Delonix regia leaf extract and its catalytic activity for nitro-aromatics hydrogenation. Ind Eng Chem Res. 2013;52:18131–18139. doi:10.1021/ie403410z
97. Mukherjee P, Roy M, Mandal BP, et al. Green synthesis of highly stabilized nanocrystalline silver particles by a non-pathogenic and agriculturally important fungus T. asperellum. Nanotechnology. 2008;19(7):75103. doi:10.1088/0957-4484/19/7/075103
98. Ali K, Ahmed B, Dwivedi S, Saquib Q, Al-Khedhairy AA, Musarrat J. Microwave accelerated green synthesis of stable silver nanoparticles with Eucalyptus globulus leaf extract and their antibacterial and antibiofilm activity on clinical isolates. PLoS One. 2015;10(7):e0131178. doi:10.1371/journal.pone.0131178
99. Wang T, Jin X, Chen Z, Megharaj M, Naidu R. Green synthesis of Fe nanoparticles using eucalyptus leaf extracts for treatment of eutrophic wastewater. Sci Total Environ. 2014;466–467:210–213. doi:10.1016/j.scitotenv.2013.07.022
100. Mehta BK, Chhajlani M, Shrivastava BD. Green synthesis of silver nanoparticles and their characterization by XRD. In: Journal of Physics: Conference Series. Vol. 836. IOP Publishing; 2017:12050.
101. Ottoni CA, Simões MF, Fernandes S, et al. Screening of filamentous fungi for antimicrobial silver nanoparticles synthesis. AMB Express. 2017;7(1):1–10. doi:10.1186/s13568-017-0332-2
102. Bagad M, Khan ZA. Poly(n-butylcyanoacrylate) nanoparticles for oral delivery of quercetin: preparation, characterization, and pharmacokinetics and biodistribution studies in Wistar rats. Int J Nanomedicine. 2015;10:3921–3935. doi:10.2147/IJN.S80706
103. Guidelli EJ, Ramos AP, Zaniquelli MED, Baffa O. Green synthesis of colloidal silver nanoparticles using natural rubber latex extracted from Hevea brasiliensis. Spectrochim Acta. 2011;82:140–145. doi:10.1016/j.saa.2011.07.024
104. Renuka R, Devi KR, Sivakami M, Thilagavathi T, Uthrakumar R, Kaviyarasu K. Biosynthesis of silver nanoparticles using Phyllanthus emblica fruit extract for antimicrobial application. Biocatal Agric Biotechnol. 2020;24:101567. doi:10.1016/j.bcab.2020.101567
105. Durán N, Marcato PD, Alves OL, De Souza GIH, Esposito E. Mechanistic aspects of biosynthesis of silver nanoparticles by several Fusarium oxysporum strains. J Nanobiotechnol. 2005;3(1):1–7. doi:10.1186/1477-3155-3-8
106. Osonga FJ, Akgul A, Yazgan I, et al. Size and shape-dependent antimicrobial activities of silver and gold nanoparticles: a model study as potential fungicides. Molecules. 2020;25(11):2682. doi:10.3390/molecules25112682
107. Rai H, Gupta RK. Biogenic fabrication, characterization, and assessment of antibacterial activity of silver nanoparticles of a high altitude Himalayan lichen-Cladonia rangiferina (L.) Weber ex FH Wigg. Trop Plant Res. 2019;6:293–298. doi:10.22271/tpr.2019.v6.i2.037
108. Romero-Urbina DG, Lara HH, Velázquez-Salazar JJ, et al. Ultrastructural changes in methicillin-resistant Staphylococcus aureus induced by positively charged silver nanoparticles. Beilstein J Nanotechnol. 2015;6(1):2396–2405. doi:10.3762/bjnano.6.246
109. He Y, Ingudam S, Reed S, Gehring A, Strobaugh TP, Irwin P. Study on the mechanism of antibacterial action of magnesium oxide nanoparticles against foodborne pathogens. J Nanobiotechnol. 2016;14(1):1–9. doi:10.1186/s12951-016-0202-0
110. Soliman H, Elsayed A, Dyaa A. Antimicrobial activity of silver nanoparticles biosynthesised by Rhodotorula sp. strain ATL72. Egypt J Basic Appl Sci. 2018;5(3):228–233.
111. Wang Y, Jett SD, Crum J, Schanze KS, Chi EY, Whitten DG. Understanding the dark and light-enhanced bactericidal action of cationic conjugated polyelectrolytes and oligomers. Langmuir. 2013;29(2):781–792. doi:10.1021/la3044889
112. Gogoi SK, Gopinath P, Paul A, Ramesh A, Ghosh SS, Chattopadhyay A. Green fluorescent protein-expressing Escherichia c oli as a model system for investigating the antimicrobial activities of silver nanoparticles. Langmuir. 2006;22(22):9322–9328. doi:10.1021/la060661v
113. Velusamy P, Kumar GV, Jeyanthi V, Das J, Pachaiappan R. Bio-inspired green nanoparticles: synthesis, mechanism, and antibacterial application. Toxicol Res. 2016;32(2):95–102. doi:10.5487/TR.2016.32.2.095
114. Veras DL, de Souza Lopes AC, Vaz da Silva G, et al. Ultrastructural changes in clinical and microbiota isolates of Klebsiella pneumoniae carriers of genes blaSHV, blaTEM, blaCTX-M, or blaKPC when subject to β--lactam antibiotics. Sci World J. 2015;2015:572128. doi:10.1155/2015/572128
115. DeLoney CR, Schiller NL. Competition of various β-lactam antibiotics for the major penicillin-binding proteins of Helicobacter pylori: antibacterial activity and effects on bacterial morphology. Antimicrob Agents Chemother. 1999;43(11):2702–2709. doi:10.1128/AAC.43.11.2702
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.