Back to Journals » International Journal of General Medicine » Volume 14
SNX20 Expression Correlates with Immune Cell Infiltration and Can Predict Prognosis in Lung Adenocarcinoma
Authors Wu GJ, Ren K, He M, Xu JX, Li ZQ, Bo D, Xue Q
Received 13 September 2021
Accepted for publication 20 October 2021
Published 3 November 2021 Volume 2021:14 Pages 7599—7611
DOI https://doi.org/10.2147/IJGM.S337198
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Gu Jie Wu,1,2,* Kuan Ren,1,2,* Min He,1,2 Jian Xun Xu,1,2 Zhen Qing Li,1,2 Ding Bo,1,2 Qun Xue3
1Research Center of Clinical Medicine, Affiliated Hospital of Nantong University, Nantong, 226001, Jiangsu, People’s Republic of China; 2Medical College of Nantong University, Nantong, 226001, Jiangsu, People’s Republic of China; 3Department of Thoracic Surgery, Affiliated Hospital of Nantong University, Nantong, 226001, Jiangsu Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qun Xue
Department of Thoracic Surgery, Affiliated Hospital of Nantong University, Nantong, 226001, Jiangsu Province, People’s Republic of China
Email [email protected]
Background: Sorting nexin-20 (SNX20) is a member of the sorting nexin family of proteins. It plays a crucial role in the regulation of innate immunity. However, the prognostic risk, potential mechanisms, immunotherapy, and other functions of SNX20 in lung adenocarcinoma (LUAD) remain unclear.
Methods: We analyzed and validated the expression and prognostic role of SNX20 in LUAD through a combination of The Cancer Genome Atlas, Gene Expression Omnibus, Oncomine, TIMER, and Human Protein Atlas databases. Further, we analyzed the correlation between SNX20 expression and clinical characteristics of LUAD, and the prognostic value of SNX20 in LUAD was evaluated. Using fitted SNX20 expression and other clinical parameters, a predictive model with predictive performance for the overall survival of patients with LUAD was constructed. The potential biological function of SNX20 in LUAD was explored using gene set enrichment analysis. In addition, we analyzed the correlation between SNX20 expression and the immune microenvironment and survival.
Results: SNX20 was downregulated in most cancer types, was associated with poor prognosis in LUAD and could be an independent prognostic factor for patients with LUAD. The predictive model developed by us had good predictive power for determining the overall survival of patients with LUAD. Biofunctional analysis revealed that genes co-expressed with SNX20 mainly promoted the immune process and inhibited the cell proliferation process in LUAD. We observed that high expression of SNX20 was accompanied by a better immune microenvironment and survival in patients with LUAD. Furthermore, the LUAD immune response was elevated with an increase in SNX20 expression. Finally, we found that SNX20 expression was significantly associated with various tumor-infiltrating immune cells, and it was widely involved in regulating various immune molecules in LUAD and affecting immune infiltration in the tumor microenvironment.
Conclusion: Our results suggested that SNX20 is a potential immune-related biomarker and therapeutic target associated with the prognosis of patients with LUAD. This provided a new strategy for the development of immunotherapeutic and prognostic markers in LUAD.
Keywords: SNX20, biomarkers, lung adenocarcinoma, prognosis, immune infiltration
Introduction
Lung cancer is still one of the leading causes of cancer-related deaths globally.1 Lung adenocarcinoma (LUAD), as the most recurring histological subtype among them, accounts for more than 40% of all lung cancer cases.2 Although a combination of cancer-related treatment techniques including surgical resection, chemotherapy, and radiotherapy has improved clinical outcomes in LUAD over the past few years, the 5-year survival rate remains below 20%. This is mainly because approximately 80% of patients with lung cancer are already at an advanced stage at the time of initial diagnosis.3 Individual treatment decisions for patients with LUAD are currently determined based on patient- and cancer-specific factors, such as tumor lymph node metastasis (TNM) stage and grade of differentiation.4 In more than 60% of patients with LUAD, survival can be improved if they do not miss early stage.5 Therefore, there is an urgent need to find more accurate LUAD markers and therapeutic targets that can improve the survival of patients with LUAD.
Sorting nexin (SNX) is a family of functional proteins involved in protein sorting and transportation. They are aberrantly expressed in a wide range of tumors and are related to cancer occurrence, metastasis, and immunity.7–10 One of the members of the SNX family, SNX20 is identified to be involved in improving the immune response activity of immune cells.11 Furthermore, it is reported that SNX20 is closely associated with the level of immune infiltration in several types of cancer. Particularly in LUAD, SNX20 expression could be a promising predictor for making therapeutic decisions regarding PD-1 inhibitors and for the assessment of therapeutic response.12 Although immunotherapy using SNX20 is promising, such studies are scarce. Therefore, the assessment of SNX20 as an immune-related biomarker in LUAD and its potential use in prognosis remains to be explored.
In this study, we obtained and analyzed a large number of tumor samples from several large databases and analyzed the expression of SNX20 in several tumor types by comparing them with normal tissues. Subsequently, we studied the correlation of SNX20 expression with clinical factors. Prognostic factors were analyzed using Cox proportional risk regression models. In addition, we further created bar graphs to predict the prognosis of patients with LUAD. Finally, we revealed the underlying mechanism of SNX20 in the development and progression of LUAD in the context of the tumor immune microenvironment.
This study elucidated the crucial role of SNX20 as a prognostic biomarker in the diagnosis and development of LUAD and revealed the potential mechanisms related to SNX20 and tumor immune microenvironment. This may lay the foundation for future clinical studies and immune-targeted therapies in LUAD.
Materials and Methods
Data Collection
Transcriptomic data and corresponding clinical information were collected from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) for 59 paraneoplastic samples and 535 LUAD samples. Detailed data of GSE72094 were collected from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds) for further validation.
Oncomine Database
Oncomine is a comprehensive analysis of gene expression and clinical relevance aggregating multiple cancer types and datasets (https://oncomine.org/resource/login.html).13 We validated the mRNA expression of SNX20 in LUAD using data from previous studies in this database.
Human Protein Atlas (HPA) Database
Human Protein Atlas (HPA; http://www.proteinatlas.org/) is a data resource website integrating atlases of tissues, cells, pathology, brain, blood, and metabolism. We analyzed the immunocytochemistry results of SNX20 from the HPA database.
TIMER
TIMER (https://cistrome.shinyapps.io/timer/) is a database that uses RNA-Seq expression profiling data to detect the relationship between gene expression and immune cell infiltration and prognostic relevance in tumor tissues.
Linkedomics Database
Differentially expressed genes associated with SNX20 are screened online through the LinkedOmics database (http://www.linkedomics.org/login.php). Not only that, this database allows GO enrichment analysis online, including cellular composition (CC), molecular function (MF) and biological process (BP). Then, we performed KEGG pathway analysis as well as Reactome analysis.
TISIDB Database
TISIDB (http://cis.hku.hk/TISIDB) is a powerful website containing a large amount of tumor immune-related data that facilitates a comprehensive study of tumor-immune interactions. The site has documented 988 genes associated with antitumor immunity by collating 4176 records from 2530 publications. Associations between genes and immune functions (eg, lymphocytes, immunomodulators, and chemokines) were also precomputed for 30 TCGA cancer types. Here, we comprehensively explored the immune system relationship of SNX20 with multiple tumors.
Statistical Analysis
SNX20 mRNA level distribution was displayed in TIMER as a box plot, using the Wilcoxon test to assess the statistical significance of differential expression. Survival curves with HR and P values or Cox P values were generated. Correlations of SNX20 expression with other genes or immune infiltration levels in certain cancer types were evaluated using Spearman correlation and statistical significance. Cutoff points were set at the median. p ≤ 0.05 was considered significant.
Results
SNX20 mRNA Levels in Various Types of Human Cancers
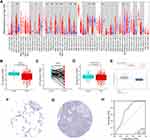
First, we assessed the role of SNX20 in tumorigenesis and analyzed the different mRNA levels of SNX20 in tumors and adjacent normal tissues in all tumors in TCGA (Figure 1A). We found that SNX20 mRNA level was significantly elevated in BRCA (breast invasive carcinoma), CHOL (cholangiocarcinoma), ESCA (esophageal carcinoma), GBM (glioblastoma multiforme), HNSC (head and neck squamous cell carcinoma), HNSC-HPV+, KIRC (renal clear cell carcinoma), KIRP (renal papillary cell carcinoma), and STAD (gastric adenocarcinoma); however, it was significantly lower in LUAD (lung adenocarcinoma), LUSC (lung squamous cell carcinoma), PAAD (pancreatic adenocarcinoma), and SKCM (cutaneous melanoma) compared with adjacent normal tissues. Analysis based on 535 tumor samples and 59 paraneoplastic samples from the TCGA database versus 288 normal samples from the GTEx database showed that SNX20 mRNA levels were lower in LUAD than in non-cancerous tissues (Figure 1B–D). Moreover, the data from Selamat et al in the Oncomine database confirmed this analysis (Figure 1E). Furthermore, to assess the protein expression of SNX20, we analyzed the results of SNX20 immunocytochemistry from the HPA database. We compared the results with the SNX20 gene expression data from the TCGA database, and the results of the analysis were consistent. These findings suggested that SNX20 was downregulated in LUAD (Figure 1F and G). Considering its prognostic value in LUAD, we generated ROC curves using normal and LUAD data from TCGA to further analyze the diagnostic value of SNX20 in LUAD. Figure 1H shows that the area under the curve (AUC) was 0.715, indicating that SNX20 had the diagnostic ability to distinguish LUAD from normal controls. These results suggested that SNX20 may play an essential regulatory role in the progression of LUAD and can be a diagnostic biomarker of LUAD.
Prognostic Value of SNX20 in Cancer Prognosis
Subsequently, we explored the prognostic potential of SNX20 in multiple cancer types. As shown in Figure 2A and B, in LUAD (HR = 0.68[0.51–0.91], P = 0.01), CESC (HR = 0.45[0.28–0.73], P = 0.001), HNSC (HR = 0.74[0. 57–0.97], P = 0.029), SARC (HR = 0.59[0.40–0.89], P = 0.011), SKCM (HR = 0.53[0.40–0.69], P < 0.001), THYM (HR = 0.09[0.01–0.71], P = 0.023), and UCEC (HR = 0.65[0.43–0.99], P = 0.043). In contrast, increased expression of SNX20 was associated with a worse prognosis in GBM (HR = 1.43[1.02–2.01], P = 0.04). To further validate the significance of SNX20 in terms of survival in patients with LUAD, we performed an analysis using LUAD data from GEO72094. The results of the analysis were consistent with the data from TCGA (HR = 0.68[0.47–0.98], P = 0.04) (Figure 2B), confirming that SNX20 expression affected the prognosis of patients with LUAD. In addition, we performed an analysis of the DSS to avoid bias due to non-cancer events (Figure 2C and D). The results were similar to the OS analysis, indicating that in LUAD (HR=0.66[0.46–0.96], P=0.028), CESC (HR=0.33[0.19–0. 60], P<0.001), HNSC (HR=0.66[0.47–0.94], P=0.022), and SARC (HR=0.61[0.39–0.95], P=0.03), SKCM (HR=0.50[0.38–0.67], P<0.001), and MESO (HR=0.50[0.26–0.94], P=0.033). In contrast, in GBM, higher SNX20 expression was associated with poorer DSS (HR = 1.44[1.00–2.07], P = 0.048). These results indicated that SNX20 expression status was significantly associated with the prognosis of patients with multiple cancer types, particularly LUAD.
Association Between SNX20 Expression and Clinicopathological Parameters
To reveal the relevance of SNX20 in the progression of LUAD, we analyzed SNX20 expression levels and clinical-pathological characters. Our data were collected from gene expression and clinical data of 535 patients with LUAD from TCGA (Table 1). Subsequently, Wilcox analysis was performed to assess the SNX20 overexpression in different clinical characters (Figure 3A–J). SNX20 overexpression was significantly associated with T stage, TNM stage, gender, age, smoking history, and immunotherapy response. Furthermore, to analyze the prognostic contribution of SNX20 expression in LUAD, we developed a Cox proportional hazards regression model. Based on the median level of SNX20 expression, all patients with LUAD were divided into a high- and low-SNX20-expression group. Univariate analysis showed that low SNX20 expression was associated with poorer late T-, N-, M-, and TNM stages and overall survival time (Figure 3K). Moreover, multivariate analysis was used to verify the predictive value of SNX20 in LUAD. The results confirmed that not only SNX20 expression but also T- and N stages were independently associated with overall survival in patients with LUAD (Figure 3L). This indicated that SNX20 expression, T stage, and N stage can be involved in guiding clinical efforts.
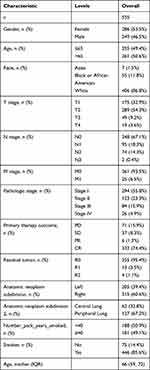 |
Table 1 Grouping of Clinicopathological Features According to SNX20 Expression Levels. |
Constructing a Prognostic Model Regarding SNX20
We constructed a prediction model of overall survival using SNX20 and independent clinical risk factors to predict the prognosis of SNX20 patients. The results of multivariate analysis indicated that SNX20 was an independent prognostic factor for LUAD. Then, we built a nomogram to use SNX20 as a biomarker for LUAD by fitting SNX20 expression and other clinical parameters (Figure 4A).In this nomogram, we used a point scale to score these variables and recorded the scores of these variables cumulatively as a total score. The probability that a LUAD patient will survive at 1, 3, and 5 years was determined by drawing a vertical line directly down from the total score axis to the outcome axis (Figure 4B).The analysis of 1000 bootstrap samples showed a C-index of 0.672 (0.652–0.693) for SNX20, indicating the accuracy of the model in terms of predictive efficiency.Therefore, we finally concluded that the nomogram is a better model than individual prognostic factors in predicting the short-term or long-term survival of LUAD patients.
Co-Expression Network of SNX20 in LUAD
To further understand the biological functions of SNX20 in LUAD, we first exposed the co-expressed genes of SNX20 in LUAD using the TCGA data in LinkedOmics. As shown in Figure 5A, 10,743 and 9245 genes were positively and negatively associated with SNX20, respectively (Supplementary Table 1). The top 50 genes positively and negatively associated with SNX20 in LUAD are shown in Figure 5B and C. GO terminology annotation showed that genes co-expressed with SNX20 mainly promoted immune processes such as interleukin production, respiratory burst, interferon-γ production, viral regulation of defense responses, and T cell activation. They inhibited cell proliferation processes in cancer such as base excision repair, DNA strand extension, histone mRNA metabolic processes, and microtubule cytoskeleton organization involved in mitosis (Figure 5D). KEGG pathway analysis revealed that co-expressed genes of SNX20 promoted graft rejection, primary immunodeficiency, graft-versus-host disease, inflammatory bowel disease, Staphylococcus aureus infection, NF-κB signaling pathway, B-cell receptor signaling pathway, Fc epsilon RI signaling pathway, inhibition of base excision repair, amino acid biosynthesis, DNA replication, nucleotide excision repair, RNA polymerase, and other aspects of enrichment (Figure 5E). Similarly, Reactome analysis confirmed that SNX20 expression promoted autoimmune responses and inhibited tumor cell proliferation during cancer development (Figure 5F). These results showed that the expression network of SNX20 could influence prognosis in LUAD by regulating tumor cell proliferation and autoimmunity.
Immune Infiltration Analysis of SNX20 in LUAD
Tumor-infiltrating lymphocytes (TILs) are infiltrating lymphocytes isolated from tumor tissue and have been identified to predict sentinel lymph node status and survival in various cancers.14 We found a significant correlation between SNX20 expression and the level of immune infiltration in LUAD after analysis using the TIMER database. As shown in Figure 6A, SNX20 expression was significantly negatively correlated with tumor purity and positively correlated with infiltration of B cells, CD8+ T cells, CD4+ cells, macrophages, neutrophils, and dendritic cells. The impact of immune infiltration in patients with LUAD on their clinical prognosis was further evaluated (Figure 6B). High levels of infiltration of B- and dendritic cells were shown to be associated with poor prognosis in patients with LUAD with overall survival of fewer than 24 months (P < 0.001 and 0.048, respectively). These findings suggested that SNX20 expression may affect LUAD and clinical prognosis by regulating the level of tumor-infiltrating immune cells. In addition, the analysis of the correlation between SNX20 and 24 immune cell infiltrates in LUAD was further expanded using TCGA data (Figure 6C and Supplementary Table 2). These cells included neutrophils; mast cells; macrophages; eosinophils; B cells; and several T cell subsets such as T effector memory, T central memory, T follicular helper, T gamma delta, Th1 cells, Th2 cells, Th17 cells, Treg, immature, activated and plasma cells; three dendritic cells (DC) types; and natural killer cells of two subtypes CD56dim and CD56bright. The results showed a significant positive correlation between SNX20 and almost all immune cell infiltration, particularly with T cell expression (Figure 6D). These results suggested that SNX20 expression affected the level of infiltration of various immune cells in LUAD. Moreover, our analysis revealed a significant association between SNX20 and Immunescore in LUAD datasets in TCGA (R = 0.910, P < 2.2e-16) and GEO (R = 0.440, P < 2.2e-16) (Figure 6E and F). In addition, patients with high immune scores had better overall survival than those with low immune scores (Figure 6G and H), which was consistent with the previous prognostic results of SNX20 expression. These data suggested that high SNX20 expression may indicate high immune cell infiltration and a better prognosis in patients with LUAD.
Relationship Between SNX20 and Immune Molecules
Finally, to gain a better understanding of the association between SNX20 and the immune microenvironment, we analyzed the correlation between SNX20 expression and various immune features. We analyzed the alterations in SNX20 expression levels and various immune characteristics. By analyzing the data in the TISIDB database, we obtained associations between SNX20 expression and various immune features. We found that high SNX20 expression resulted in a significant increase in the infiltration of TILs in most cancers. In Figure 7A, we have listed the top 6 TILs with the highest correlation, with MDSC_abundance having the highest correlation with SNX20 (R = 0.853, P < 2.2e-16). Figure 7B shows the correlation between SNX20 and various cancer immunosuppressants. In LUAD, SNX20 showed a strong positive correlation with the top 6 immunosuppressants, especially CSF1R (R = 0.824, P < 2.2e-16). Additionally, some common immunosuppressants such as PD-1 and PD-L1 showed a significant positive correlation. Figure 7C shows the correlation between SNX20 expression and immunostimulants. High expression of SNX20 stimulated stronger immune responses, and the correlation with the top 6 immunostimulants was more significant than 0.72. Figure 7D shows the correlation between SNX20 expression and MHC molecules; we could see that high SNX20 expression elicited a strong immune response to MHC in various tumors. Figure 7E shows the correlation between SNX20 expression and chemokines, where SNX20 showed the highest correlation with CCL19 (R = 0.654). Figure 7F shows the correlation between SNX20 expression and receptors; similar to the previous results of other immune profiles, SNX20 not only correlated more than 0.82 with CCR2 and CCR5, but also showed a strong positive correlation with other receptors.
Thus, our study demonstrated that SNX20, as a novel prognostic marker, was widely involved in the regulation of various immune molecules in LUAD and influenced the immune infiltration in the tumor microenvironment (TME).
Discussion
LUAD is known to be a highly malignant and aggressive disease among various cancers, with more than 30% of early-stage patients likely to face recurrence or develop metastases.15,16 Due to the absence of obvious symptoms, patients often miss the best time for treatment. Therefore, more accurate potential biomarkers of LUAD are urgently needed to predict prognosis and guide individualized patient treatment.
The SNX family of proteins is closely associated with cancer. As a member of SNX, SNX20, like other members, has been identified to be involved in many critical cellular activities, including protein endocytosis, sorting, transport, degradation, and signaling.6 Additionally, it has been shown that SNX1 sorts EGFR into lysosomes for degradation through a biological pathway. In this way, it is engaged in EGFR resistance in lung cancer.17 Moreover, SNX10, which has reduced expression in colorectal cancer, promotes tumor formation and proliferation.18 In addition to its ability to promote tumor development, the role of SNX20 in immune promotion is even more pronounced because SNX20 expression enhances the immune response activity of immune cells and the effect of PD-1 inhibitors and is strongly associated with the level of immune infiltration in 30 cancer types, particularly in LUAD.11,12 Therefore, SNX20 could provide new insights into the diagnosis and treatment of patients with LUAD by acting as a potential prognostic biomarker and target for immunotherapy.
Tumorigenesis is a multistep, multilevel process in which regulation of the TME plays a crucial role.19 The TME is generally composed of three components: extracellular matrix (ECM), stromal cells, and immune cells.20 It has been shown that the ECM provides support for the growth of tumor cells and can promote the progression of cancer cells, such as growth and migration, in concert with stromal cells.21 Immune cells tend to exhibit immunosuppression in TMB, and the regulation of this mechanism plays a crucial role in the prognosis and immunotherapy in patients with cancer22. For example, macrophages often turn into tumor-associated macrophages that drive tumor progression, invasion, and metastasis.23 Additionally, tumor-infiltrating dendritic cells tend to contribute to immunosuppression and tolerance.24 Thus, in most cases, the dominant role of TME is immunosuppression, blocking antitumor immunity, and maintaining tumor progression. In recent years, immunotherapy has gradually gained attention and has emerged as a promising therapeutic modality for treating various cancers, particularly LUAD.25 This has led to increasing recognition of the importance of TME. Immune cell infiltration by TME has been shown to correlate with immunotherapy efficacy and survival of patients with solid tumors.26 Therefore, the exploration of immune cell infiltration could reveal more insights into the key factors affecting the survival of patients with LUAD.
In this study, we analyzed bioinformatics data from a large number of patients with LUAD to explore the potential role of the novel prognostic marker SNX20 in LUAD. Transcriptomic analysis of clinical samples, including several large databases, revealed that SNX20 mRNA levels were remarkably lower in both overall samples and matched samples from individual patients than in non-cancerous lung tissues. Furthermore, SNX20 expression levels remarkably influenced clinical factors, including primary tumor status and tumor pathological stage. Our analysis showed that SNX20 expression was involved in the pathological progression of LUAD tissues and was an independent factor determining the prognosis of patients with LUAD. Thus, as a potential prognostic marker, further clinical validation of SNX20 should be performed. Finally, we combined SNX20 with other critical clinical factors (T-stage, N-stage, and SNX20 expression levels) to comprehensively assess the line graphs. Our model was constructed based on the complementary perspective of individual tumors. It provided individualized scores for individual patients. According to calibration chart validation, our model exhibited an excellent agreement between actual and forecasted values of OS at 1, 3, and 5 years for patients with LUAD. Thus, our model constructed with SNX20 for the prediction of survival of patients with LUAD will likely be a valuable new prognostic approach for clinicians.
Biological function analysis of genes co-expressed with SNX20 in LUAD confirmed that high SNX20 expression improved the prognosis in LUAD mainly by inhibiting tumor progression and regulating autoimmune-related pathways. To explore the underlying mechanisms, we explored the correlation between SNX20 expression, immune cell infiltration, immune microenvironment, and prognosis. By analyzing the immunological and survival data of patients with LUAD from the TIMER database, we found that SNX20 expression levels were highly positively correlated with the infiltration levels of B cells, CD4+ and CD8+ T cells, dendritic cells, macrophages, and neutrophils, and high infiltration of B cells and dendritic cells led to a better prognosis of patients with LUAD. This suggested that high expression of SNX20 could improve the prognosis of patients with LUAD by enhancing immune cell infiltration. To verify this result, we performed a combined validation analysis using data from TCGA, GEO, and TISIDB. Similar to previous results, we found that high expression of SNX20 induced an increase in the majority of immune cell infiltrates, particularly T cells. Many studies have confirmed that T cells are the main enforcers of tumor progression; they act by mediating immune responses, including inhibition of T cell trafficking to the tumor and interference with antigen-presenting cells and effector T cells.21,26 In addition, SNX20 is widely involved in regulating various immune molecules including tumor-specific TILs in LUAD to influence immune infiltration in the TME. In addition, TILs play an important role in tumor control and the prevention of tumor progression.27 These results were consistent with previous biofunctional analyses. Subsequently, based on the ESTIMATE algorithm, we found a significant positive correlation between SNX20 expression and immune scores in patients with LUAD in TCGA and GEO databases. Patients with high SNX20 expression or high immune scores had longer survival. These results indicated that high SNX20 expression can improve the immune microenvironment in tumors of patients with LUAD by increasing the level of immune cell infiltration, thus improving the prognosis of patients. Furthermore, our study revealed that SNX20 has a great impact on immune cell infiltration in TME of most cancers. Therefore, an in-depth study on the potential role of SNX20 expression in the field of tumor immunotherapy is warranted.
In summary, we concluded that a strong association existed between SNX20 as a novel prognostic marker and immune infiltration in patients with LUAD. This study constructed an overall survival prediction model with some accuracy using SNX20 and independent clinical risk factors. Not only that, we found that elevated SNX20 expression levels were significantly associated with increased levels of immune infiltration in LUAD. Moreover, high SNX20 expression resulted in a better immune microenvironment and prognosis in patients with LUAD. Further analysis revealed that SNX20 was widely involved in regulating immune molecules in various cancers, particularly LUAD, and influencing immune infiltration in the TME. In future studies, we can try to assess the malignancy and predict the prognosis of patients by measuring the expression of SNX20 in surgical specimens of patients with LUAD. Additionally, we can assess the immune microenvironment status and develop immunotherapeutic agents targeting SNX20. As a high potential immunotherapeutic marker for LUAD, we strongly recommend researchers in the field of tumor immunology to further investigate the role of SNX20 in various tumors, particularly in LUAD, and to deeply explore the biological functions of SNX20 in the immune microenvironment and prognosis of patients with LUAD. This will be beneficial for LUAD immunotherapy.
Ethics Approval
TCGA and GEO belong to public databases. The patients involved in the database have obtained ethical approval. Users can download relevant data for free for research and publish relevant articles. Our study is based on open source data, so there are no ethical issues and other conflicts of interest.The Ethics Committee of Affiliated Hospital of Nantong University has granted exemptions from approval for research related to the use of such public databases.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi:10.3322/caac.21387
2. Shi J, Hua X, Zhu B, et al. Somatic genomics and clinical features of lung adenocarcinoma: a retrospective study. PLoS Med. 2016;13(12):e1002162. doi:10.1371/journal.pmed.1002162
3. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi:10.3322/caac.21349
4. Politi K, Herbst RS. Lung cancer in the era of precision medicine. Clin Cancer Res. 2015;21:2213–2220. doi:10.1158/1078-0432.CCR-14-2748
5. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):
6. Cullen PJ. Endosomal sorting and signalling: an emerging role for sorting nexins. Nat Rev Mol Cell Biol. 2008;9(7):574–582. doi:10.1038/nrm2427
7. You Y, Zhou C, Li D, et al. Sorting nexin 10 acting as a novel regulator of macrophage polarization mediates inflammatory response in experimental mouse colitis. Sci Rep. 2016;6:20630. doi:10.1038/srep20630
8. Bendris N, Williams KC, Reis CR, et al. SNX9 promotes metastasis by enhancing cancer cell invasion via differential regulation of RhoGTPases. Mol Biol Cell. 2016;27(9):1409–1419. doi:10.1091/mbc.E16-02-0101
9. Chen X, Chen S, Hang W, Huang H, Ma H. MiR-95 induces proliferation and chemo- or radioresistance through directly targeting sorting nexin1 (SNX1) in non-small cell lung cancer. Biomed Pharmacother. 2014;68(5):589–595. doi:10.1016/j.biopha.2014.04.008
10. Zhou Q, Huang T, Jiang Z, et al. Upregulation of SNX5 predicts poor prognosis and promotes hepatocellular carcinoma progression by modulating the EGFR-ERK1/2 signaling pathway. Oncogene. 2020;39(10):2140–2155.
11. Schaff UY, Shih HH, Lorenz M, et al. SLIC-1/sorting nexin 20: a novel sorting nexin that directs subcellular distribution of PSGL-1. Eur J Immunol. 2008;38(2):550–564. doi:10.1002/eji.200737777
12. Fan L, Li L, Huang C, Huang S, Deng J, Xiong J. Increased SNX20 and PD-L1 levels can predict the clinical response to PD-1 inhibitors in lung adenocarcinoma. Onco Targets Ther. 2020;13:10075–10085. doi:10.2147/OTT.S262909
13. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi:10.1593/neo.07112
14. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi:10.1158/0008-5472.can-17-0307
15. Ettinger DS, Akerley W, Bepler G, et al. Non-small cell lung cancer. J Natl Compr Canc Netw. 2010;8:740–801. doi:10.6004/jnccn.2010.0056
16. Wu CF, Fu JY, Yeh CJ, et al. Recurrence risk factors analysis for stage I non-small cell lung cancer. Medicine (Baltimore). 2015;94:e1337. doi:10.1097/MD.0000000000001337
17. Nishimura Y, Takiguchi S, Yoshioka K, Nakabeppu Y, Itoh K. Silencing of SNX1 by siRNA stimulates the ligand-induced endocytosis of EGFR and increases EGFR phosphorylation in gefitinib-resistant human lung cancer cell lines. Int J Oncol. 2012;41(4):1520–1530. doi:10.3892/ijo.2012.1578
18. Zhang S, Yang Z, Bao W, et al. SNX10 (sorting nexin 10) inhibits colorectal cancer initiation and progression by controlling autophagic degradation of SRC. Autophagy. 2020;16(4):735–749.
19. Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting tumor microenvironment for cancer therapy. Int J Mol Sci. 2019;20(4):840. doi:10.3390/ijms20040840
20. Oliver AJ, Lau P, Unsworth AS, et al. Tissue‐dependent tumor microenvironments and their impact on immunotherapy responses. Front Immunol. 2018;9:70. doi:10.3389/fimmu.2018.00070
21. Buoncervello M, Gabriele L, Toschi E. The janus face of tumor microenvironment targeted by immunotherapy. Int J Mol Sci. 2019;20(17):4320. doi:10.3390/ijms20174320
22. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi:10.1038/nm.3394
23. Ngambenjawong C, Gustafson HH, Pun SH. Progress in tumor‐associated macrophage (TAM)‐targeted therapeutics. Adv Drug Deliv Rev. 2017;114:
24. Pitt JM, Marabelle A, Eggermont A, Soria JC, Kroemer G, Zitvogel L. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol. 2016;27(8):
25. Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):
26. Kalinski P, Talmadge JE. Tumor immuno‐environment in cancer progression and therapy. Adv Exp Med Biol. 2017;1036:
27. Simoni Y, Becht E, Fehlings M, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557(7706):575–579. doi:10.1038/s41586-018-0130-2
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.