Back to Journals » Cancer Management and Research » Volume 12
SNHG7 Contributes to the Progression of Non-Small-Cell Lung Cancer via the SNHG7/miR-181a-5p/E2F7 Axis
Authors Wang L, Zhang L, Wang L
Received 3 December 2019
Accepted for publication 16 April 2020
Published 7 May 2020 Volume 2020:12 Pages 3211—3222
DOI https://doi.org/10.2147/CMAR.S240964
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Liming Wang,1 Lili Zhang,2 Liwei Wang3
1Department of Interventional, Shandong Provincial Chest Hospital, Jinan, Shandong, People’s Republic of China; 2Thoracoscopic Ward, Shandong Provincial Chest Hospital, Jinan, Shandong, People’s Republic of China; 3Department of Radiology, Tianbao Township Health Center, Taian, Shandong, People’s Republic of China
Correspondence: Liwei Wang
Department of Radiology, Tianbao Township Health Center, Tianbao Town, High-Tech Zone, Taian City, Shandong Province, People’s Republic of China
Email [email protected]
Background: Non-small-cell lung cancer (NSCLC) is a common malignant tumor with very high mortality. Small nucleolar RNA host gene 7 (SNHG7) was associated with many tumors progression. We aimed to explore the role and regulatory mechanism of SNHG7 in the development of NSCLC.
Methods: The expression of SNHG7, miR-181a-5p and E2F transcription factor 7 (E2F7) was detected by quantitative real-time polymerase chain reaction (qRT-PCR). The protein expression of E2F7 was evaluated by Western blot. Cell Counting Kit-8 (CCK-8) assay was conducted to explore cell proliferation. Flow cytometry was used to examine cell apoptosis. The clonogenic examination was performed to reflect cell population dependence and proliferative ability. Transwell assay was used to assess cell migration and invasion. The potential target relationship between miR-181a-5p and SNHG7 or E2F7 was analyzed by dual-luciferase reporter assay. A xenograft mouse model was generated to verify the effect of SNHG7 on tumor growth in vivo.
Results: SNHG7 and E2F7 were increased, while miR-181a-5p was decreased in NSCLC. Knockdown of SNHG7 suppressed cell viability, clonogenic, migration, invasion and tumor growth, and promoted cell apoptosis. SNHG7 acted as a sponge of miR-181a-5p and E2F7 was directly interacted with miR-181a-5p. Overexpression of miR-181a-5p had the same functional effect as SNHG7 knockdown on the progression of NSCLC cells. E2F7 was negatively correlated with miR-181a-5p and positively correlated with SNHG7. Moreover, miR-181a-5p inhibition or E2F7 overexpression abolished the effect of SNHG7 knockdown on the progression of NSCLC cells.
Conclusion: SNHG7 regulated the development of NSCLC cells by the miR-181a-5p/E2F7 axis.
Keywords: SNHG7, miR-181a-5p, E2F7, NSCLC
Introduction
Lung cancer is the most commonly diagnosed cancer in the world. Statistical analysis of 185 countries and 36 tumors (besides all cancers united) by the Global Cancer Observatory at 2018 displayed that lung cancer has become the highest mortality-related disease in human due to 18.4% of the mortality rate in the total cancer death, closely followed by breast cancer (11.6%) and colorectal cancer (10.2%).1,2 Non-small-cell lung cancer (NSCLC) occupies about 80–85% of all lung cancer cases.3,4 Patients are often diagnosed as NSCLC in the middle and late stages; current treatments for NSCLC are limited to surgery, radiotherapy and chemotherapy. To find more effective therapeutic methods, more and more scientists have focused on the analysis of long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) to expound the relying molecular signaling pathways. At present, many lncRNAs and miRNAs are associated with diverse cellular processes and cancer progression. For example, lncRNA small nuclear RNA host gene1 (MALAT1) was noticeably upregulated in NSCLC cells and inhibited cell viability, proliferation, migration and invasion by interacting with miR-145-5p and regulating MTDH.5 MiR-5702 inhibited cell proliferation and invasion and encouraged apoptosis in A549 cells by means of suppressing ZEB1 expression.6
Small nucleolar RNA host gene 7 (SNHG7) is known as a novel lncRNA with 2176 bp in length (NR_003672). Plentiful studies publicized that SNHG7, as an oncogene, was mainly upregulated in multiple tumors and participated in the regulation of the development of tumors by various different adjustment pathways. For instance, SNHG7 interference inhibited cell proliferation and apoptosis in gastric cancer cells,7 bladder cancer cells,8 and thyroid cancer cells.9 Through epithelial-to-mesenchymal transition (EMT) initiation and the Notch-1 pathway, SNHG7 knockdown significantly inhibited MCF-7 cell proliferation, invasion and tumor growth in mice by sponging miR-34a in breast cancer.10 About the relationship between SNHG7 and NSCLC, She et al found that SNHG7 supported proliferation, anti-apoptosis and metastasis in NSCLC cells by antagonizing miR-193b and inducing FAIM2.11 Nevertheless, the mechanism of SNHG7 on regulating the progression of NSCLC cells is still insufficiently understood.
MiRNAs have also been one of the focuses on cancer researches in recent years. As a course of endogenous RNAs with 18–25 nucleotides (nts) in length, miRNAs can bind with 3ʹUTR of target mRNAs and lead to mRNAs degradation by convening lncRNAs. Accumulating documents exposed that deregulation of miRNAs linked with tumorigenesis and the progression of multiple tumors, including miR-181a-5p. For instance, ZEB1-AS1 promoted colorectal cancer cell proliferation and NSCLC cell invasion and angiogenesis in Wnt/β-catenin signaling by sponging miR-181a-5p.11–13 Upregulation of miR-181a-5p accelerated cell proliferation and invasion, and repressed apoptosis in cervical cancer cells (HeLa and SiHa) by targeting inositol polyphosphate-5-phosphatase A (INPP5A).14 Ectopic expression of miR-181a-5p promoted cell proliferation, colony formation, cell cycle transition, invasion, metastasis and EMT of gastric cancer.15 In short, miR-181a-5p plays a crucial role in the development of tumors, but there is little evidence on the role of miRNA-181a-5p in the regulation of lung cancer cell motility and development.
E2F transcription factor 7 (E2F7) belongs to the E2F/DP family winged-helix DNA-binding domain with 5135 bp in length (XM_011537966). Abundant evidence has revealed that E2F7 abnormality plays an important role in the development of cancer cells. Overexpression of E2F7 was correlated with a high risk of relapse and poor prognosis through inhibiting miR-15a/16 cluster by competing with E2F1 in breast cancer patients who received tamoxifen treatment.16 The gain of E2F7 function neutralized the impacts of miR-30a-5p on cell proliferation and metastasis.17 About the effect of E2F7 on NSCLC cells, Wang et al reported that E2F7 was upregulated in NSCLC tissues and associated with poor prognosis and suppressed cell proliferation, migration, invasion, tumor growth, the EMT and AKT pathways in NSCLC cells through targeting miR-935,18 but the relevant research is still scarce.
In the present research, the interaction relationship among SNHG7, miR-181a-5p and E2F7 was firstly verified. SNHG7 was overexpressed in NSCLC cells and tissues, and positively correlated with E2F7, but negatively correlated with miR-181a-5p. SNHG7, as a competing endogenous RNA and overview of miR-181a-5p, impeded cell proliferation, colony formation, invasion, migration and tumor growth, and promoted cell apoptosis by regulating E2F7 expression. Our results provided new insights into the molecular meaning of SNHG7 in the progression of NSCLC cells and new potential targets for clinical application.
Methods
Patient Samples and Cells Acquisition
Our study was approved by the Ethics Committee of Shandong Provincial Chest Hospital, and we also obtained the written informed consents from all patients underwent the surgical resection in Shandong Provincial Chest Hospital, thirty pairs of lung carcinoma tissue samples were collected from lung cancer patients. Human lung cancer cell lines (NCI-H520, SPC-A1 and H-23) and normal histiocytes (BEAS-2B) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were cultivated in Roswell Park Memorial Institute (RPMI) 1640 (Gibco, Rockville, MD, USA) with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in an incubator containing 5% CO2.
Total RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted using TRIzol reagent kit (Thermo Fisher Scientific). EasyScript First-Strand cDNA synthesis superMix (TransGen Biotech, Beijing, China) or miRNA 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) was used to synthesize cDNA. QRT-PCR was performed using AceQ Universal SYBR qPCR Master Mix (Vazyme) The melting curves of SNHG7 miR-181a-5p and E2F7 were visualized on Applied Biosystems (ABI) 7500 Real-Time PCR System (Thermo Fisher Scientific), the relative expression levels were analyzed on the basis of the calculation model of 2−∆∆Ct manner. Additionally, U6 or GAPDH was attended as the endogenous control of miRNA and mRNA, separately. The synthesized primers sequences were as follows: SNHG7: 5ʹ- GTCAGCCGCATCTTCTTTTG-3ʹ and 5ʹ-GCGCCCAATACGACCAAATC-3ʹ; E2F7: 5ʹ-AGGGATGGAGGTAAATTGTTTAACACT-3ʹ and 5ʹ-TTTCCCCATCTTCAACTGCAA-3ʹ; miR-181a-5p: 5ʹ-GCGAACATTCAACGCTGTCGGTGAGT-3ʹ and 5ʹ- CAGTGCGTGTCGTGGAGT-3ʹ; GAPDH: 5ʹ-TCTCCTCTGACTTCAACAGC-3ʹ and 5ʹ -CCACCCTGTTGCTGTAGCCAA −3ʹ; U6: 5ʹ -TCGCTTCGGCAGCACA-3ʹ and 5ʹ-ACGCTTCACGAATTTGCGT-3ʹ. All primers were synthesized from Ribobio (Guangzhou, China).
Cell Transfection
SNHG7 shRNA (sh-SNHG7) and negative control (sh-NC), miR-181a-5p mimics (miR-181a-5p) and its control (miR-NC), miR-181a-5p inhibitors and its control (anti-NC), the overexpression vector of E2F7 (E2F7) and its control (Vector) were synthesized from GenePharma (Shanghai, China). Lipofectamine 2000 reagent (Invitrogen, Waltham, MA, USA) was used to assist in transfection for the following experiments.
Cell Viability Assay
Cell viability was conducted by Cell Counting Kit-8 (CCK-8) experiment. Cells at the logarithmic phase were digested and blown into the single-cell by 0.25% trypsin, then cells were suspended in RPMI1640 medium (Gibco) containing 10% FBS (Thermo Fisher Scientific), and injected into 96-well plates at a density of 2.0×103 cells per well and cultured in an incubator containing 5% CO2 at 37°C until adhering to the wall. The cells were added 20 μL CCK-8 (Beyotime Corporation, Shanghai, China) for 2 h at 37°C. Titertek Multiskan Ascent Labsystems 354 Plate Reader (Madison, Wisconsin, USA) was used to measure cell viability at the designated time points (0 h, 24 h, 48 h, 72 h).
Clonogenic Test
Cells were suspended in RPMI1640 medium (Gibco) with 10% FBS (Thermo Fisher Scientific), and cultured at 37°C in a humid incubator containing 5% CO2 for 2 weeks until clonal colonies visible to the naked eye. After being washed twice by phosphate-buffered saline buffer (PBS), the cells were fixed with pure methyl alcohol and stained with Giemsa (Solarbio, Beijing, China) for 30 min. The excess dye was cleared out, and the plates were inverted on a transparent film with the cartridge. Low power microscopy was used to count the number of cells. The clone formation rate was calculated following calculation model: The clonogenic rate (%) = (number of clones/number of vaccinations cells) × 100%.
Cell Apoptosis Assay
AnnexinV-fluorescein isothiocyanate (BD Pharmingen, San Diego, CA, USA) and propidium iodide (PI; BD Pharmingen) were used to detect cell apoptosis. Cells at logarithmic phase were washed two times with PBS and blown off with 400 μL 1× binding buffer suspension and diluted to the concentration of 1.0×106/mL. Afterward, 5 μL AnnexinV-fluorescein isothiocyanate (BD Pharmingen) and 10 μL PI (BD Pharmingen) were added to stain the cells at 4°C in the dark. After 1 h, the apoptosis rate was analyzed on the flow cytometry.
Cell Migration Assay
Cells were injected into the upper chamber at a density of 1.0×106 cells per well, while the lower chamber was overspread with 700 μL of medium (Gibco) with 10% FBS (Thermo Fisher Scientific). The chambers were continuously cultivated for 24 h. The upper chamber was alone taken out and immersed into 800 μL methanol for 30 min, then stained with Giemsa (Solarbio). With neutral drying, the areas were randomly selected and calculated under the microscope, and the observed results were statistically analyzed.
Cell Invasion Assay
Cells were plated into the upper chamber with matrigel, the lower chamber was added with 500 μL medium (Gibco) including 10% FBS (Thermo Fisher Scientific). Transwells were grown together at 37°C for 1 d, then washed two times with PBS and fixed with 5% glutaraldehyde at 4°C, then stained with Giemsa (Solarbio). Cotton swabs were used to remove the non-invasive cells. The calculated areas were randomly selected under the microscope, and the results were statistically analyzed.
Dual-Luciferase Reporter Assay
StarBase v2.0 and TargetScan 7.2 were used to predict the prospective binding sites among SNHG7, miR-181a-5p and E2F7. To confirm the relationship between SNHG7 and miR-181a-5p, SNHG7-wild type (wt) or mutant (mut) was co-transfected into NCI-H520 and SPC-A1 cells with miR-181a-5p or its negative control (mi-NC); To investigate the connection between miR-181a-5p and E2F7, E2F7 types (E2F7 3ʹUTR-wt/E2F7 3ʹUTR-mut) with miR-181a-5p or miR-NC were co-transfected into NCI-H520 and SPC-A1 cells. The specific steps should be strictly followed by instructions of the Dual Luciferase Reporter Gene Assay Kit (Yeasen, Shanghai, China). The luciferase activity was imagined on a Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA).
Protein Expression Detection
Protein samples were isolated by Protein Expression Lysate Control Kit (Thermo Fisher Scientific) and separated by 10% SDS-PAGE (Mlbio, Shanghai, China). Subsequently, the protein was transferred to nitrocellulose membranes (GE Healthcare, Westborough, MA, USA). The membranes were blocked with 5% skim milk powder, and cultivated with primary antibody at 4°C overnight. After washed twice by Tris-buffered saline and tween-20 (TBST), the membranes were incubated with the horseradish peroxidase (HRP)-conjugated secondary antibody for 2 h. The destination strips were imagined on the ECL detection system (Thermo Fisher Scientific).
Xenograft Mouse Model
The research was agreed by the Animal Protection Association of Shandong Provincial Chest Hospital. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. Experimental procedures were approved by the Institutional and Local Committee on the Care and Use of Animals of Shandong Provincial Chest Hospital. All animals received humane care according to the National Institutes of Health (USA) guidelines. Lentiviral vector for stable SNHG7 shRNA (sh-SNHG7) and the negative control (sh-NC) were purchased from Genechem (Shanghai, China) and six-week-old athymic nude mice were bought from Shanghai Experimental Animal Center (Shanghai, China). Lung cancer cells were transfected with sh-SNHG7 or sh-NC and cultured at a concentration of 2×106/mL. Transfected cells were subcutaneously injected into the left-axilla of nude mice (n=5 per group). Tumor volume was measured once every 5 days, and its growth curve was established. After 36 days, tumors were collected and tumor weight was measured.
Statistical Analysis
The difference was evaluated by the Student’s t-test or One-Way ANOVA on software SPSS 18.0. All values were included at least three independent duplications for each group of trials and presented as mean ± standard deviation (SD), P < 0.05 was represented statistically significant.
Results
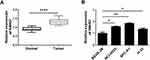
SNHG7 Was Upregulated in NSCLC Tissues and Cells
To detect the expression of SNHG7 in lung cancer tissues and cells, thirty pairs of lung carcinoma tissue samples and adjacent normal histiocytes were collected to extract total RNA for quantitative real-time PCR. The results suggested that SNHG7 was notably upregulated in lung cancer tissues compared with adjacent normal tissues (Figure 1A). In addition, qRT-PCR was conducted to determine the expression of SNHG7 in human lung cancer cell lines (NCI-H520, SPC-A1 and H-23) and the relative normal cells (BEAS-2B). The data indicated that the expression level of SNHG7 was notably increased in NCI-H520, SPC-A1 and H-23 cells compared with BEAS-2B cells (Figure 1B). The expression profile of SNHG7 implied that SNHG7 might play an important role in the progression of NSCLC.
Knockdown of SNHG7 Inhibited the Development of NSCLC Cells
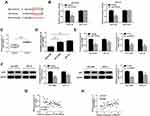
To investigate the function of SNHG7 on the development of NSCLC cells, NCI-H520 and SPC-A1 cells were transfected with sh-SNHG7 or the negative control (sh-NC) for a series of functional investigations. The data of qRT-PCR (Figure 2A) showed that compared with NCI-H520 and SPC-A1 cells transfected with sh-NC, the expression of SNHG7 was decreased in sh-SNHG7 transfected NCI-H520 and SPC-A1 cells. CCK-8 and clonogenic assays (Figure 2B and C) revealed that knockdown of SNHG7 reduced cell viability and clone formation rate. However, flow cytometry analysis (Figure 2D) detected that the apoptosis rate was raised after SNHG7 knockdown in NCI-H520 and SPC-A1 cells. Transwell test indicated that the number of cell migration (Figure 2E) and invasion (Figure 2F) were clearly reduced in NCI-H520 and SPC-A1 cells transfected with sh-SNHG7. Moreover, we obtained a successful knockdown efficiency of sh-SNHG7-s1 in both NCI-H520 and SPC-A1 cells (Supplement Figure 1A). Knockdown of SNHG7 could significantly inhibit cell proliferation and decrease the number of colonies in NSCLC cells (Supplement Figure 1B and C). Moreover, SNHG7 deletion enhanced the rate of apoptosis in NSCLC cells (Supplement Figure 1D). Transwell assays showed that knockdown of SNHG7 dramatically suppressed cell migration and invasion in both NCI-H520 and SPC-A1 cells (Supplement Figure 1E and F). These data demonstrated that SNHG7 could inhibit the progress of NSCLC cells through suppressing cell viability, clonogenic, migration and invasion, and promoting cell apoptosis.
SNHG7 Knockdown Negatively Regulated miR-181a-5p Expression by Direct Interaction
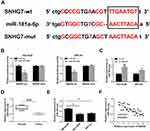
To indicate the connection between SNHG7 and miR-181a-5p in NSCLC cells, starBase v2.0 predicted the presumed binding sequences between SNHG7 and miR-181a-5p (Figure 3A). Dual-luciferase reporter assay was used to determine the target relationship between SNHG7 and miR-181a-5p. The results exhibited that the luciferase activity in NCI-H520 and SPC-A1 cells co-transfected with miR-181a-5p and SNHG7-wt were markedly inhibited compared to miR-NC and SNHG7-wt co-transfected cells, but there was no change in SNHG7-mut group (Figure 3B). The data of qRT-PCR showed that the expression of miR-181a-5p was highly expressed in sh-SNHG7 transfected NCI-H520 and SPC-A1 cells compared with sh-NC transfected cells (Figure 3C). As expected, miR-181a-5p was downregulated in lung cancer tissues and cells (Figure 3D and E), and the correlation index was −0.7711 between the expression of SNHG7 and miR-181a-5p, signifying that miR-181a-5p and SNHG7 were negatively correlated in tumor tissues (Figure 3F). These results proved SNHG7 might play an important role in lung cancer cells by sponging miR-181a-5p.
MiR-181a-5p Inhibited the Progression of NSCLC Cells
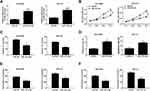
To investigate the effect of miR-181a-5p on lung cancer cells, miR-181a-5p or miR-NC was transfected into NCI-H520 and SPC-A1 cells. QRT-PCR was used to detect the transfection efficiency, and a series of functional verification tests were carried out, including cell viability, clonogenic, migration, invasion and apoptosis assays. The results displayed that miR-181a-5p was upregulated in NCI-H520 and SPC-A1 cells transfected with miR-181a-5p (Figure 4A). Cell proliferation (Figure 4B) was reduced in NCI-H520 and SPC-A1 cells following the overexpression of miR-181a-5p, and the number of clonogenic (Figure 4C), migration (Figure 4E) and invasion (Figure 4F) were also declined in NCI-H520 and SPC-A1 cells after miR-181a-5p upregulation. Moreover, the apoptosis rate of NCI-H520 and SPC-A1 cells transfected with miR-181a-5p was ascended compared to miR-NC group (Figure 4D). These data confirmed that miR-181a-5p participated in the progression of lung cancer cells.
SNHG7 Positively Regulated E2F7 Expression via Sponging miR-181a-5p in NSCLC Cells
TargetScan 7.2 predicted that E2F7 was a target gene of miR-181a-5p and the presumed binding sites were exhibited in Figure 5A. Dual-luciferase reporter assay exhibited that miR-181a-5p and E2F7-wt co-transfection reduced the luciferase activity in NCI-H520 and SPC-A1 cells compared with miR-NC and E2F7-wt co-transfected group, while the luciferase activity was not affected in E2F7-mut group (Figure 5B). To identify the relationships among E2F7, miR-181a-5p and SNHG7, E2F7 mRNA and protein expression was detected by qRT-PCR and Western blot, respectively. E2F7 mRNA was elevated in NSCLC tissues and cells compared with normal tissues and BESA-2B cells (Figure 5C and D). E2F7 was downregulated in NCI-H520 and SPC-A1 cells transfected with miR-181a-5p and NCI-H520 and SPC-A1 cells transfected with sh-SNHG7 (Figure 5E). Additionally, Western blot validated the expression of E2F7 protein was significantly declined in NCI-H520 and SPC-A1 cells transfected with miR-181a-5p or sh-SNHG7 (Figure 5F). The correlation between E2F7 and SNHG7 or miR-181a-5p was analyzed in lung cancer tissues, as shown in Figure 5G and H, E2F7 was negatively correlated with miR-181a-5p but positively correlated SNHG7 in NSCLC tissues. These findings suggested E2F7 was positively regulated by SNHG7 via miR-181a-5p in NSCLC cells.
MiR-181a-5p Inhibition or E2F7 Overexpression Reversed the Effects of SNHG7 Knockdown on Cell Proliferation, Clonogenic, Migration, Invasion and Apoptosis in NSCLC Cells
To research the relationship among SNHG7 miR-181a-5p and E2F7,NCI-H520 and SPC-A1 cells were co-transfected with sh-NC, sh-SNHG7, sh-SNHG7+miR-181a-5p inhibitors, sh-SNHG7+anti-NC, sh-SNHG7+E2F7, or sh-SNHG7+ vector. Firstly, the expression profiles of E2F7 mRNA and protein were checked by qRT-PCR and Western blot, respectively. E2F7 mRNA (Figure 6A) and protein (Figure 6B and C) levels were all decreased in NCI-H520 and SPC-A1 cells transfected with sh-SNHG7but miR-181a-5p inhibitors or E2F7 transfection restored the decrease. To further verify the association among SNHG7, miR-181a-5p and E2F7 in regulating the progression of NSCLC, CCK-8 assay, clonogenic assay, flow cytometry analysis and transwell assay were conducted. CCK-8 assay (Figure 6D) showed that the cell viability was significantly degraded in NCI-H520 and SPC-A1 cells transfected with sh-SNHG7, but miR-181a-5p inhibitors or E2F7 transfection overturned the effect. Moreover, SNHG7 silencing suppressed colony formation (Figure 6E), cell migration (Figure 6G) and cell invasion (Figure 6H), and facilitated cell apoptosis in NCI-H520 and SPC-A1 cells (Figure 6F), whereas all these influences were abolished by the downregulation of miR-181a-5p or the upregulation of E2F7. These data indicated SNHG7 regulated cell proliferation, clonogenic, migration, invasion and apoptosis in NSCLC cells via the miR-181a-5p/E2F7 axis.
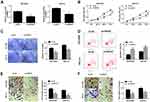
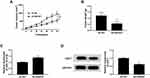
Silencing of SNHG7 Suppressed Tumor Growth in vivo
To investigate the role of SNHG7 in the progression of NSCLC, a xenograft tumor mouse model was established via injecting sh-SNHG7 transfected SPC-A1 cells into the mice. Then tumor volume and weight were measured and we found that tumor volume and weight were markedly decreased in sh-SNHG7 group compared to sh-NC group (Figure 7A and B). Afterward, the levels of miR-181a-5p and E2F7 in the collected tumor samples were determined by qRT-PCR and Western blot assay, respectively. The data displayed that miR-181a-5p was elevated and E2F7 was reduced in the tumors of sh-SNHG7 group (Figure 7C and D). Collectively, SNHG7 knockdown could suppress tumorigenesis in vivo.
Conclusion
Through the application of biological monitoring technology, we found that SNHG7 and E2F7 were upregulated in NSCLC tissues and cells, while miR-181a-5p had opposite expression pattern. Knockdown of SNHG7 suppressed NSCLC cell viability, clonogenic, migration, invasion and promoted cell apoptosis in vitro and suppressed tumor growth in vivo. From the results of online biological prediction and dual-luciferase reporter assay, we verified that SNHG7 directly interacted with miR-181a-5p, and E2F7 was a downstream target gene of miR-181a-5p. MiR-181a-5p inhibition or E2F7 overexpression counteracted the consequence of SNHG7 knockdown on the progression of NCI-H520 and SPC-A1 cells.
In recent years, people have tended to search for new therapeutic targets about cancer progression through the pathway of interaction between lncRNAs and miRNAs. SNHG7 was found to act as an oncogene in various tumors and modulated the expression and function of mRNA by interacting with miRNAs. In colorectal cancer, SNHG7 was significantly upregulated and positively regulated GALNT1 level through sponging miR-216b during the tumor progression.19 In glioblastoma (GBM), SNHG7 expression was significantly increased in GBM tissues and cell lines, downregulation of SNHG7 remarkably crushed cell proliferation, migration and invasion and induced cell apoptosis in GBM cell lines (A172 and U87) through sponging miR-5095 and concomitantly activating the Wnt/β-catenin signaling pathway.20 SNHG7 overexpression was faithfully connected with the poor prognosis in prostate cancer cells and demonstrated that SNHG7 markedly suppressed cell proliferation and cycle progression, and induced cell cycle arrest at G0/G1 phase and suppressed tumor growth through miR-503/Cyclin D1 pathway.21 Moreover, SNHG7 was also found to play important role in NSCLC by targeting miRNAs. Pang and his colleagues indicated that SNHG7 exerted promotion effects on cell growth and metastasis by sponging miR-449a to regulate TGIF2 expression in NSCLC.22 SNHG7 was involved in the tumor progression of NSCLC via miR-193b/FAIM2 axis.11 These results were consistent with our findings.
Here, the interaction relationship among SNHG7, miR-181a-5p and E2F7 was firstly reported, we mainly found that SNHG7 was increased in NSCLC tissues and cells, SNHG7 interference suppressed cell proliferation, clonogenic, migration, invasion and promoted cell apoptosis in vitro, and restrained tumor growth in vivo by the SNHG7/miR-181a-5p/E2F7 axis. Undoubtedly, miR-181a-5p inhibitor or E2F7 overexpression could repair the effect of SNHG7 on NSCLC development. Our results would provide new potential targets for clinical application and novel insights into the molecular meaning of SNHG7 in the progression of NSCLC cells. However, due to the lack of tissue sample size and pathway-related validation, we were unable to define the pathway accurately, and need further experimental verification.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. YiRen H, YingCong Y, Sunwu Y, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16(1):174. doi:10.1186/s12943-017-0743-3
3. Li S, Ma Y, Hou X, et al. MiR-185 acts as a tumor suppressor by targeting AKT1 in non-small cell lung cancer cells. Int J Clin Exp Pathol. 2015;8(9):11854–11862.
4. Song L, Peng L, Hua S, et al. miR-144-5p enhances the radiosensitivity of non-small-cell lung cancer cells via targeting ATF2. BioMed Res. 2018;2018:5109497. doi:10.1155/2018/5109497
5. Lu Q, Shan S, Li Y, Zhu D, Jin W, Ren T. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J. 2018;32(7):3957–3967. doi:10.1096/fj.201701237RR
6. Zhang C, Xue Q, Xu Z, Lu C. MiR-5702 suppresses proliferation and invasion in non-small-cell lung cancer cells via posttranscriptional suppression of ZEB1. J Biochem Mol Toxicol. 2018;32(7):e22163. doi:10.1002/jbt.22163
7. Wang MW, Liu J, Liu Q, et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21(20):4613–4622.
8. Zhong X, Long Z, Wu S, Xiao M, Hu W. LncRNA-SNHG7 regulates proliferation, apoptosis and invasion of bladder cancer cells assurance guidelines. J BUON. 2018;23(3):776–781.
9. Wang YH, Huo BL, Li C, Ma G, Cao W. Knockdown of long noncoding RNA SNHG7 inhibits the proliferation and promotes apoptosis of thyroid cancer cells by downregulating BDNF. Eur Rev Med Pharmacol Sci. 2019;23(11):4815–4821. doi:10.26355/eurrev_201906_18067
10. Sun X, Huang T, Liu Z, Sun M, Luo S. LncRNA SNHG7 contributes to tumorigenesis and progression in breast cancer by interacting with miR-34a through EMT initiation and the Notch-1 pathway. Eur J Pharmacol. 2019;856:
11. She K, Yan H, Huang J, Zhou H, He J. miR-193b availability is antagonized by LncRNA-SNHG7 for FAIM2-induced tumour progression in non-small cell lung cancer. Cell Prolif. 2018;51(1). doi:10.1111/cpr.12406
12. L L, Wang Q, Zou M-L, He X-A, Lv -J-J. Overexpressed lncRNA ZEB1-AS1 promotes cell invasion and angiogen esis through Wnt/beta-catenin signaling in non-small cell lung cancer. Int J Clin Exp Pathol. 2017.
13. Lv SY, Shan TD, Pan XT, et al. The lncRNA ZEB1-AS1 sponges miR-181a-5p to promote colorectal cancer cell proliferation by regulating Wnt/beta-catenin signaling. Cell Cycle. 2018;17(10):1245–1254. doi:10.1080/15384101.2018.1471317
14. Yang M, Zhai X, Ge T, Yang C, Lou G. miR-181a-5p promotes proliferation and invasion and inhibits apoptosis of cervical cancer cells via regulating inositol polyphosphate-5-phosphatase A (INPP5A). Oncol Res. 2018;26(5):703–712. doi:10.3727/096504017x14982569377511
15. Mi Y, Zhang D, Jiang W, et al. miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signalling activation. Cancer Lett. 2017;389:
16. Chu J. E2F7 overexpression leads to tamoxifen resistance in breast cancer cells by competing with E2F1 at miR-15a16 promoter.pdf. Oncotarget. 2015. doi:10.18632/oncotarget.5128
17. Ye YY, Mei JW, Xiang SS, et al. MicroRNA-30a-5p inhibits gallbladder cancer cell proliferation, migration and metastasis by targeting E2F7. Cell Death Dis. 2018;9(3):410. doi:10.1038/s41419-018-0444-x
18. Wang C, Li S, Xu J, Niu W, Li S. microRNA-935 is reduced in non-small cell lung cancer tissue, is linked to poor outcome, and acts on signal transduction mediator E2F7 and the AKT pathway. Br J Biomed Sci. 2019;76(1):17–23. doi:10.1080/09674845.2018.1520066
19. Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Discov. 2018;9(7):722. doi:10.1038/s41419-018-0759-7
20. Ren J, Yang Y, Xue J, et al. Long noncoding RNA SNHG7 promotes the progression and growth of glioblastoma via inhibition of miR-5095. Biochem Biophys Res Commun. 2018;496(2):712–718. doi:10.1016/j.bbrc.2018.01.109
21. Qi H, Wen B, Wu Q, et al. Long noncoding RNA SNHG7 accelerates prostate cancer proliferation and cycle progression through cyclin D1 by sponging miR-503. Biomed Pharmacother. 2018;102:
22. Pang L, Cheng Y, Zou S, et al. Long noncoding RNA SNHG7 contributes to cell proliferation, migration, invasion and epithelial to mesenchymal transition in non-small cell lung cancer by regulating miR-449a/TGIF2 axis. Thoracic Cancer. 2020;11(2):264–276. doi:10.1111/1759-7714.13245
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.