Back to Journals » Cancer Management and Research » Volume 11
SNHG1 contributes to proliferation and invasion by regulating miR-382 in breast cancer
Authors Zheng S, Li M, Miao K, Xu H
Received 18 December 2018
Accepted for publication 22 April 2019
Published 18 June 2019 Volume 2019:11 Pages 5589—5598
DOI https://doi.org/10.2147/CMAR.S198624
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Shipeng Zheng, Mengquan Li, Keke Miao, Han Xu
Department of Breast Surgery, the First Affiliated Hospital of Zhengzhou University, Zhengzhou City, Henan Province 450052, China
Introduction: Long non-coding RNAs (lncRNAs) are key regulators in multiple cancers. lncRNA, SNHG1, was shown to be associated with tumorigenesis. However, little is known about the role SNHG1 plays in breast cancer. The aim of the study was to study the role and underlying mechanism of SNHG1 regulation in breast cancer.
Methods: Quantitative real-time PCR was used to measure the levels of SNHG1, miR-382 and ZEB1 levels in breast cancer tissue or cells. The proliferation, colony formation, migration and invasion of breast cancer cells, under SNHG1 knockdown achieved by transfection of SNHG1-specific siRNAs, were assessed by Cell Counting Kit-8, colony forming, scratch wound and transwell assays. Bioinformatical analysis and luciferase assay were used to explore the interaction between SNHG1 and its potential miRNA target. Western blot was used to evaluate the expression of epithelial-to-mesenchymal transition (EMT) markers. MDA-MB-231 cells with or without SNHG1 knockdown were used to initiate tumor xenografts in vivo. Tumor growth and expression of SNHG1, miR-382-5p and EMT markers were evaluated.
Results: SNHG1 upregulation was observed in breast cancer tissues and cells. Knockdown of SNHG1 attenuated breast cancer proliferation, colony formation, migration and invasion. A miRNA, miR-382-5p, was identified as the target of SNHG1. A reciprocal negative regulation was found between SNHG1 and miR-382-5p. SNHG1 knockdown attenuated EMT both in vitro and in vivo. miR-382-5p transfection reversed the tumor-promoting role by SNHG1. In vivo, SNHG1 knockdown decreased breast tumor growth.
Conclusion: SNHG1 promotes breast cancer through the regulation of miR-382-5p and EMT markers. Our results report SNHG1 as a novel miRNA that govern the progression of breast cancer, providing a potential new therapeutic target in breast cancer.
Keywords: breast cancer, therapeutic target, SNHG1, miR-382
Introduction
Breast cancer is a highly prevalent cancer and a leading cause of cancer-related death in women.1 Although the survival of patients with localized breast cancers has been significantly improved, conventional treatment methods, including surgery, chemotherapy and radiotherapy, fail to deliver a desirable therapeutic outcome for patients with advanced-stage cancer. To address this, overwhelming efforts have been devoted to the development of new targeted strategies for breast cancer.2 Exploration of the molecular basis in breast cancer tumorigenesis, progression, metastasis and recurrence is of paramount importance to develop new strategies to improve patient survival.3
An emerging class of oncogenes that regulate cancer progression are long non-coding RNAs (lncRNAs), which are of over 200 np in length, serving as molecular sponges of other regulatory molecules, including miRNAs.4 Dysregulated lncRNAs post-transcriptionally activate or suppress certain oncogenes linked to the proliferation, migration, invasion, etc. of the cancer cells. To date, a handful of cancer-regulatory lncRNAs have been discovered in breast cancer, including, ANCR,5 ATB,6 UCA,7 NEAT1,8 MEG3,9 etc, whereby the lncRNAs may adopt cancer-promoting or cancer-suppressing roles. As such, strategies targeted to these cancer-related lncRNA are promising novel approaches to inhibit breast cancer progression.10
SNHG1 is one such tumor-promoting lncRNA proven to govern many cancers. For example, SNHG1 upregulation was found correlated to lung cancer,11 prostate cancer,12 heptacellular cancer,13,14 etc., whereby SNHG1 plays important roles in activating putative oncogenic pathways, such as Wnt/β-catenin signaling pathway. Several miRNAs, including miR-101-3p, miR-199-3p and miR-195, are demonstrated to be regulated by SNHG1.11–13 Suppression of SNHG1 was shown as an efficacious approach to attenuate cancer progression.11,14 However, the role of SNHG1 in breast cancer is rarely studied.15 Moreover, how SNHG1 involves in the putative driving force of cancer progression, epithelial-to-mesenchymal transition (EMT), which takes a pivotal part in the metastasis of breast cancer,7 remains largely unknown.
Herein, we strive to elucidate the function and target of SNHG1 in breast cancer and test the therapeutic effects of SNHG1 knockdown in cancer progression. Here, we identified a target of SNHG1 in breast cancer, miR-382-5p, which has been previously reported as a tumor-suppressive miRNA.16 It was shown that SNHG1 upregulation is a hallmark of breast cancer cell proliferation, migration and invasion and SNHG1 knockdown effectively attenuated breast cancer aggressiveness both in vitro and in vivo. We also showed that miR-382-5p serves as an essential mediator of the tumor-promoting effects of SNHG1. The results of the study could shed light on the potential of this lncRNA as a therapeutic target in breast cancer to improve breast cancer diagnosis and therapy.
Materials and methods
Clinical tissue specimens
A total of 52 pairs of breast cancer tissues and paired non-tumor tissues were collected from the First Affiliated Hospital of Zhengzhou University from June 2012 to September 2014. The study was approved by the First Affiliated Hospital of Zhengzhou University Ethics Committee and all patients provided written informed consent. Tissue samples were snap-frozen in liquid nitrogen and stored at −80°C until use. All the trials were conducted in accordance with the Declaration of Helsinki.
Cell lines and culture
Human normal human breast epithelial cell (MCF-10a) and breast cancer cell lines (MCF-7, MDA-MB-231, ZR-75-30 and MDA-MB-453) were purchased from ATCC (Manassas, VA, USA). RPMI 1640 medium supplemented with 10% fetal bovine serum was used to culture the cells at 37°C.
Oligonucleotides transfection
Oligonucleotides, including siRNA against SNHG1 (Si-SNHG1), short-hairpin RNA plasmid directly targeting SNHG1 (sh-SNHG1), miR-382-5p inhibitor, miR-382-5p mimics and their controls, were c synthesized by GenePharma (Shanghai, China). Transfection of the oligonucleotide into cells was conducted using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The sequence of siRNA for SNHG1 and control was Si-SNHG1, sense 5′-CCCCATGATGGTTCCTCAGTT-3′, antisense 3′-GGAAAGCAAGTGCAGGTTAGTC-5′; control: sense: 5ʹ - GGCCGTCACTCAATGATTCCG −3ʹ, antisence: 5ʹ -UUTTGGATGGCATACGCATGA −3ʹ.
BLAST alignment
NCBI’s BLAST suite was used to perform alignment searches. The top search result with an e-value <0.01 was reported. RNA transcripts were allowed to have multiple exons aligned to different non-contiguous regions of a chromosome.
Quantitative real-time PCR (qRT-PCR)
The miRNeasy Mini Kit (Qiagen, Valencia, CA, USA) was used to extract total RNA from tissues and cells in accordance with the manufacturer’s instructions. Concentration and quality of RNA were evaluated with NanoDrop 2000 (Thermo Fisher, Wilmington, DE, USA). TransScript first-strand cDNA synthesis SuperMix (TransGen, Beijing, China) was used to synthesize cDNA. RT-PCR was conducted using SYBR green qPCR SuperMix (Applied Biosystems Life Technologies, Foster, CA, USA) in ABI prism 7500 system (Applied Biosystems Life Technologies). Fold changes of each gene were calculated by 2−ΔΔCt (cycle threshold). Expression levels of miRNA and lncRNA/target gene were normalized by U6 and GADPH, respectively.The following sequences were used in this study: SNHG1, forward: sense 5′- CCCCATGATGGTTCCTCAGTT-3′, reverse 3′-GGAAAGCAAGTGCAGGTTAGTC-5′; miR-382-5p, forward: 5′- TAAAATAGGAGTACTGTCTAA-3′, reverse, 5′- ATTAGTAAATTGGCTGCTGCAG-3’; GAPDH, forward, 5′-TCGACAGTCA GCCGCATCTTCTTT-3′, reverse, 5′-ACCAAATCCGTTGACTCCGACCTT-3′
Luciferase reporter assay
Oligonucleotides that encode SNHG1 cDNA encompassing microRNA-binding sites are inserted into the pmirGLO plasmids (Promega, Madison, WI, USA). Site-directed mutagenesis PCR was used to generate mutant SNHG1 (pmirGLO-SNHG1-MUT) with platinum pfx DNA polymerase, which served as a negative control. Luciferase reporter plasmids and target miR-382-5p mimics or miR-NC mimics were co-transfected into cells by Lipofectamine 2000. After 48 hrs, relative luciferase activity was assessed in a luminometer by Dual-Luciferase Reporter Assay System (Promega).
Cell proliferation assay
Cell Counting Kit-8 (CCK-8; Dojindo, JPN) and colony formation assay were used to monitor cell proliferation. In CCK-8 assay, cells were harvested and seeded into 96-well plates. After 24, 48, 72 or 96 hrs, CCK-8 assay agent of 10 μL was added to each well. After incubation for 2 hrs, DMSO was added and the absorbance of the solution was measured using an enzyme immunoassay analyzer (Bio-rad, Hercules, CA, USA). Colony formation assay stable transfected cells were placed into six-well plates and maintained in media for 2 weeks. The medium was replaced every 4 days. Colonies were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich, St Louis, MO, USA) for 15 mins. The visible colonies were then counted. All experiments were conducted three times in triplicate.
Cell migration and invasion assay
Wound healing assay and transwell assay were used to assess cell migration and invasion, respectively. Briefly, for wound healing assay, cells seeded in six-well plates were cultured to 90% confluence. The monolayer cells were manually scraped using a 10-uL sterile pipette tip. The wounding scratches at 24 hr was measured and expressed as the relative percentage of the initial distance at 0 hr following formula: migration rate = migration distance/original distance. For transwell assay, cells (5×104) were suspended in serum-free DMEM and added to chambers (8 mm, BD Biosciences) coated with BD BioCoat Matrigel. After incubation, the cells on the upper membrane surface were removed with a cotton tip. Then, the member was fixed and stained by violet crystalline.
Western blot analysis
RIPA buffer (Sigma-Aldrich) was used to lyze cells. The mixture was centrifuged with 12,000 g to remove insoluble components, followed by protein concentration measurement using BCA assay. Thirty micrograms of protein were separated by SDS–PAGE and transferred to PVDF membrane (Millipore, Bedford, MA, USA). PVDF membrane was blocked with non-fat milk and incubated with primary antibodies, including anti-ZEB1 (1:1,000, Abcam, Cambridge, MA), anti-GADPH (1:1,000, Abcam). After washing with TBST, HRP-conjugated goat anti-rabbit IgG (Abcam) was used to incubate the membrane at room temperature for 2 hrs. Finally, protein bands were visualized using ECL detection kit (Beyotime Biotechnology, Shanghai, China).
Lentivirus construction and infection
A recombinant lentiviral vector encoding SNHG1-shRNA was introduced into pFU-GW-RNAi vector. Cells were seeded onto six-well plates at the density of 2×105 cells per well. After 12 hrs, cells were infected with Lv-shRNA-NC or Lv-shRNA- SNHG1 at 10 MOI, respectively. Virus-containing culture medium was replenished with fresh RPMI-1640 medium 12 hrs postinfection.
In vivo experiments
Animal study was performed according to the experimental protocols and approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Zhengzhou University. All mice were housed and maintained under specific pathogen-free conditions, with free access to water and food. To initiate tumor xenografts, 1×107 MCF-7 cells were subcutaneously injected into the flank of BALB/c mice (6 weeks). Tumor growth was monitored by caliper measurements every 3 days and tumor volume was calculated according to the following formula: volume =0.5× length × width × width. The mice were sacrificed with 120 μL of 10% chloral hydrate (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) and the tumors were harvested and weighed.
Immunohistochemical staining
Tumor tissues were cryosectioned at the thickness of 5 μm. After washing by PBS, goat serum (Boster, Wuhan, China) was used to block sections for 30 mins at room temperature. Subsequently, Ki67 antibody (Bioss Antibodies, Inc, 1:200) or Colorimetric TUNEL Apoptosis Assay Kit (Beyotime, Shanghai, China) were incubated with the tissue overnight at 4°C at 37°C for 60 mins. Following this, after washing by PBS, sections were incubated with Polink-1 HRP DAB Detection System One-step polymer detection system (ZSGB-BIO, Beijing, China) for 20 mins. Slides were then counterstained with hematoxylin.
Statistical analysis
All the statistical data are presented as the means ± SD Two-tailed Student’s t-test or one-way ANOVA followed by the LSD post hoc test was performed for comparisons between groups. Expression correlation assays were analyzed using Pearson’s coefficient correlation. Differences in patient survival were performed using the Kaplan–Meier method and analyzed by log-rank test. A value of p<0.05 was considered to be statistically significant.
Results
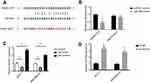
Breast cancer is characterized by SNHG1 upregualtion
To explore the role of SNHG1 in breast cancer, we first compared SNHG1 levels in breast tumor tissues and paired adjacent normal tissues from 54 patients. qRT-PCR analysis of SNHG1 levels showed a significant SNHG1 upregulation in tumor tissue compared normal tissues (p<0.05, n=54, Figure 1A). To correlate SNHG1 levels to patient survival, we grouped the patients to high-expression group (n=29) and low-expression group (n=25) based on the median level of SNHG1 expression (Figure 1B). A poorer survival in high SNHG1 expression was observed (Figure 1C, p=0.011). Therefore, breast cancer is characterized by an upregulation in SNHG1, which contributes to the poor survival of patients.
SNHG1 upregulation promotes breast cancer cell proliferation, migration and invasion
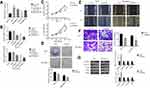
To confirm the role of SNHG1 in breast cancer, we analyzed SNHG1 levels in breast cancer cell line, including MCF-7,MDA-MB-231,ZR-75-30 and MDA-MB-453. The MCF-10a cells, which do not form tumors in vivo, were used as controls. As expected, breast cancer cells demonstrated higher SNHG1 levels compared to MCF-10a (p<0.05, Figure 2A). We next proceeded to examine whether SNHG1 upregulation is linked to a higher tumor aggressiveness. Consequently, knockdown of SNHG1 was performed through siRNAs. Three siRNAs specific to SNHG1 were designed, namely, si-SNHG1-1, si-SNHG1-2 and si-SNHG1-3, among which si-SNHG1-3 exhibited the most efficacious SNHG1 knockdown (Figure 2B), although all siRNAs were able to reduce SNHG1 expression. Because of the highest knockdown efficiency, si-SNHG1-3 was used for subsequent studies. MTT (Figure 2C), colony formation (Figure 2D), scratch wound (Figure 2E) and transwell assays (Figure 2F) were used to evaluate the effect of SNHG1 knockdown on cell proliferation, colony formation, migration and invasion, respectively, whereby a marked attenuation of those tumor aggressiveness indicators was seen in both MCF-7 and MDA-MB-231 cells. Given that EMT is closely linked to cell aggressiveness, we analyzed EMT markers, including E-cadherin, N-cadherin, vimentin and ZEB1 in MCF-7 and MDA-MB-231 cells using Western blot. As shown in Figure 2G, SNHG1 knockdown led to upregulation of E-cadherin, downregulation of N-cadherin, vimentin and ZEB1, which suggested dampened EMT. Together, these evidence indicated that SNHG1 is an important promoter of breast cancer aggressiveness.
miR-382-5p is a target of SNHG1
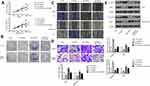
To unveil the underlying mechanism of the tumor-promoting role of SNHG1, we performed a bioinformatical analysis to search for the target of SNHG1. As shown in Figure 3A, a binding site was found between miR-382-5p and wild-type SNHG1 (SNHG1 WT). Mutagenesis was conducted within the binding site on SNHG1 to miR-382-5p, yielding SNHG1 MUT. Luciferase assay (Figure 3B) was used to validate the interaction between SNHG1 and miR-382-5p, which showed that miR-382-5p overexpression, achieved by transfection of miR-382-5p mimics, induced a prominent downregulation of SNHG1-WT-driven luciferase activity, but not the SNHG1-MUT-driven luciferase activity. In both MCF-7 and MDA-MB-231 cells, miR-382-5p mimic transfection resulted in a dramatic downregulation of SNHG1 levels, while miR-382-5p inhibitor led to a significant SNHG1 upregulation (Figure 3C). Reciprocally, si-SNHG1 transfection upregulated miR-382 expression (Figure 3D). Further, miR-382 inhibitor was shown to antagonize si-SNHG1 in breast cancer colony formation and invasion (Figure S1). Therefore, SNHG1 and miR-382 levels are negatively correlated.
miR-382 mediates the regulation of breast cancer aggressiveness by SNHG1
Considering the close correlation between miR-382-5p and SNHG1, we next evaluated whether the manipulation of miR-382-5p expression affects the tumor-promoting effects by SNHG1. To this end, MCF-7 and MDA-MB-231 were transfected with si-NC, si-SNHG1, miR-382-5p inhibitor or si-SNHG1+miR-382-5p inhibitor. It was shown that SNHG1 knockdown reduced cell proliferation, colony formation, migration and invasion; miR-382-5p inhibitor transfection exerted the opposite effects (Figure 4A–D). Interestingly, co-transfection of si-SNHG1 and miR-382-5p inhibitor resulted in cell proliferation, colony formation, migration and invasion similar to those of the cells transfected with si-NC, indicating that miR-382-5p inhibitor abolished the tumor-inhibitory effects of si-SNHG1. Evaluation of EMT markers (Figure 4E) also confirmed that the EMT-suppressing effect by si-SNHG1, as well as the EMT-enhancing effect by miR-382 inhibitor, was abrogated by co-transfection of si-SNHG1 and miR-382-5p inhibitor.
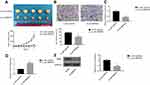
SNHG1 knockdown attenuates breast tumor growth in vivo
MDA-MB-231 cells transfected with Lv-sh-Control or Lv-sh-SNHG1 were used to initiate breast tumor xenografts in mice. Eight weeks after tumor inoculation, tumors were harvested and photographed. As shown in Figure 5A, tumors with Lv-sh-SNHG1 transfected demonstrated observably smaller sizes, which correlated well with the slower tumor growth rate. A significant reduction in Ki-67 levels was also seen in tumors with Lv-sh-SNHG1 transfection (Figure 5B and C). Meanwhile, SNHG1 downregulation and miR-382-5p upregulation were observed in tumors with Lv-sh-SNHG1 transfection (Figure 5D). The EMT marker, ZEB1, was also downregulated (Figure 5E). Hence, these data suggested that SNHG1 knockdown was an effective approach to reduce tumor growth in the studied model.
Discussions and conclusions
With the advent of newly approved gene therapy strategies,10 increasing oncogenes are being tested as therapeutic targets in cancers. Because of the potent and broad-spectrum regulation by lncRNAs, knockdown of cancer-promoting lncRNAs in cancer is a promising approach to induce efficacious therapeutic responses. In the present study, we studied SNHG1, a lncRNA proven to adopt powerful regulatory roles in lung cancer,11 prostate cancer,12 hepatocellular cancer,14 etc., with the purpose to characterize it as a therapeutic target in breast cancer. Previously, the upregulation of SNHG1 has been implicated in one study,15 but a systemic study of the role of SNHG1 in breast cancer progression is needed. We showed that, both in tissue and in cell levels, SNHG1 upregulation is a hallmark of breast cancer. Importantly, higher SNHG1 expression is associated with a poor patient survival. These findings are in agreement to previous studies, which showed that SNHG1 overexpression has been observed in many cancers.11–14 This also qualifies SNHG1 as a potential biomarker of breast cancer. Indeed, SNHG1 has already been shown of significant diagnostic potential in hepatocellular carcinoma.14 To further characterize the diagnostic potential of SNGH1, correlation between SNHG1 levels, preferably in serum, and breast cancer grade may be necessary.
The pro-oncogenic role of SNHG1 was further validated by the attenuated cell proliferation, colony formation and invasion in cells with SNHG1 knockdown, mediated by si-SNHG1 transfection. Consistently, previous evidence also supported the potent anti-cancer effects of SNHG1 knockdown.11–13 Notably, SNHG1 silencing was efficient in attenuating EMT, which presumably contributed to the anti-tumor effects. In vivo, SNHG1 knockdown also attenuated tumor growth. Our study is paralleled by several efforts in translating lncRNA-silencing strategy in therapy of cancers.17,18 Further studies are needed to evaluate whether SNHG1 knockdown would improve the survival of tumor-bearing mice, particularly in those with metastatic cancer. To foster clinical translation, an in situ delivery to tumor, possibly mediated by viral or non-viral delivery systems,19 would be desirable.
Our study showed that miR-382-5p is a target of SNHG1 in breast cancer. Prior evidence indicated that miR-382-5p is a tumor-suppressing miRNA in a number of cancers, including osteosarcoma,20 ovarian cancer21 and colorectal cancer.16 Notably, miR-382-5p was reported as a mediator of EMT,21 which concurs with our findings. Our data also clarified the role of miR-382-5p in breast cancer, underscoring miR-382-5p as an important therapeutic target in cancer. Here, we proposed that the pro-oncogenic role of SNHG1 is at least partly attributed to its regulation of miR-382-5p. Other miRNAs, such as miR-101-3p, miR-199-3p and miR-195, have also been suggested as the target of SNHG1,11–13 which likely also play important parts in breast cancer.
In summary, we clarified SNHG1 as a tumor-promoting lncRNA in breast cancer. SNHG1 silencing was able to exert anti-tumor effects both in vitro and in vivo. Concurrently, EMT was attenuated by SNHG1 silencing. Therefore, SNHG1 is a potential therapeutic target in breast cancer.
Acknowledgments
This work was supported by Henan Provincial Key R&D and Promotion Special Support Project (grant no: 182102310362) and Henan Province Higher Education Key Research Projects Guiding Plan (grant no: 19B320042).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. 68:394–424.
2. Gotwals P, Cameron S, Cipolletta D, et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat Rev Cancer. 2017;17(5):286. doi:10.1038/nrc.2017.17
3. Nagini S. Breast cancer: current molecular therapeutic targets and new players. Anticancer Agents Med Chem. 2017;17(2):152–163.
4. Peng W, Koirala P, Mo Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661. doi:10.1038/onc.2017.184
5. Li Z, Hou P, Fan D, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24(1):59. doi:10.1038/cdd.2016.95
6. Shi S-J, Wang L-J, Yu B, Li Y-H, Jin Y, Bai X-Z. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6(13):11652. doi:10.18632/oncotarget.v6i13
7. Xiao C, Wu C, Hu H. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2016;20(13):2819–2824.
8. Zhang M, Wu W, Wang Z, Wang X. lncRNA NEAT1 is closely related with progression of breast cancer via promoting proliferation and EMT. Eur Rev Med Pharmacol Sci. 2017;21:1020–1026.
9. Zhang W, Shi S, Jiang J, Li X, Lu H, Ren F. LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed Pharmacother. 2017;91:312–319. doi:10.1016/j.biopha.2017.04.085
10. Stein CA, Castanotto D. FDA-approved oligonucleotide therapies in 2017. Mol Ther. 2017;25(5):1069–1075. doi:10.1016/j.ymthe.2017.03.023
11. Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget. 2017;8(11):17785.
12. Li J, Zhang Z, Xiong L, et al. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;487(1):146–152. doi:10.1016/j.bbrc.2017.03.169
13. Zhang H, Zhou D, Ying M, et al. Expression of long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) exacerbates hepatocellular carcinoma through suppressing miR-195. Med Sci Monit. 2016;22:4820. doi:10.12659/MSM.898574
14. Zhang M, Wang W, Li T, et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–79. doi:10.1016/j.biopha.2016.02.036
15. Yu F, Bracken CP, Pillman KA, et al. p53 represses the oncogenic Sno-MiR-28 derived from a SnoRNA. PLoS One. 2015;10(6):e0129190. doi:10.1371/journal.pone.0129190
16. Zhou B, Song J, Han T, et al. MiR-382 inhibits cell growth and invasion by targeting NR2F2 in colorectal cancer. Mol Carcinog. 2016;55(12):2260–2267. doi:10.1002/mc.22466
17. Haemmerle M, Gutschner T. Long non-coding RNAs in cancer and development: where do we go from here?. Int J Mol Sci. 2015;16(1):1395–1405. doi:10.3390/ijms16011395
18. Lavorgna G, Vago R, Sarmini M, Montorsi F, Salonia A, Bellone M. Long non-coding RNAs as novel therapeutic targets in cancer. Pharmacol Res. 2016;110:131–138. doi:10.1016/j.phrs.2016.05.018
19. Ren Y, Li RQ, Cai YR, Xia T, Yang M, Xu FJ. Effective codelivery of lncRNA and pDNA by pullulan-based nanovectors for promising therapy of hepatocellular carcinoma. Adv Funct Mater. 2016;26(40):7314–7325. doi:10.1002/adfm.201603041
20. Xu M, Jin H, Xu C-X, et al. miR-382 inhibits tumor growth and enhance chemosensitivity in osteosarcoma. Oncotarget. 2014;5(19):9472. doi:10.18632/oncotarget.2418
21. Tan H, He Q, Gong G, et al. miR-382 inhibits migration and invasion by targeting ROR1 through regulating EMT in ovarian cancer. Int J Oncol. 2016;48(1):181–190. doi:10.3892/ijo.2015.3241
Supplemental materials
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.