Back to Journals » Cancer Management and Research » Volume 11
Small RNA sequencing reveals a novel tsRNA-26576 mediating tumorigenesis of breast cancer
Authors Zhou J, Wan F , Wang Y, Long J, Zhu X
Received 22 December 2018
Accepted for publication 24 March 2019
Published 2 May 2019 Volume 2019:11 Pages 3945—3956
DOI https://doi.org/10.2147/CMAR.S199281
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Jun Zhou,1 Fang Wan,1 Yike Wang,1 Jinpei Long,1 Xuan Zhu2,3
1Department of Surgery, The Women’s Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang Province, People’s Republic of China; 2The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang Province, People’s Republic of China; 3The Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education, Hangzhou, Zhejiang Province, People’s Republic of China
Purpose: As a malignancy that develops from breast tissue, breast cancer has been widely regarded as the most common type of cancer threatening the health of women worldwide. Emerging evidence has demonstrated that tsRNAs might play a vital part in the tumorigenesis and progression of several types of cancers. However, the functions of tsRNAs in breast cancer remain largely unknown. Here, we investigated the functions of tsRNA-26576 in tumorigenesis of breast cancer.
Patients and methods: In this study, the tsRNA deregulation states in breast cancer patients (four cancer tissues and four adjacent normal tissues) were evaluated using small RNA sequencing. And then, RT-PCR was used to detected the tsRNA-26576 expression level in breast cancer patients.
Results: A total of 263 tsRNAs were identified as significantly differentially expressed, of which 75 were upregulated, and 188 were downregulated. The functional classification through KEGG pathway database illustrated that the most significant pathway enriched by the targets of differentially expressed tsRNAs was the pathway in cancer. Among these differently expressed tsRNAs, we found that tsRNA-26576 was remarkably upregulated in cancer tissue in comparison with adjacent normal tissue. Meanwhile, RT-PCR results verified that tsRNA-26576 expression level was highly upregulated in 10 paired samples from breast cancer patients. Besides, tsRNA-26576 was found to motivate cellular multiplication and migration while suppressing cellular apoptosis in MDA-MB-231 cells. Moreover, mRNA sequencing results showed that several tumor suppressor genes, including FAT4 and SPEN, were upregulated after delivering tsRNA-26576 inhibitor in MDA-MB-231 cells.
Conclusion: We found tsRNA-26576 was upregulated in breast cancer tissue, and it could promote the cell growth while inhibite cell apoptosis. Therefore, tsRNA-26576 might serve as a potential clinical therapy target and a predictive marker for breast cancer.
Keywords: malignancy, tsRNA, MDA-MB-231 cells, predictive marker
Introduction
Breast cancer is known as the second most commonly diagnosed cancer in American females. With around one million new cases diagnosed annually accounting more than 400,000 death, breast cancer has become a dreadful public health problem obsessing women worldwide. In spite of the fact that the breast cancer death rate has declined 38% from peak rates on account of remarkable progress made in early detection and treatment, almost all of the patients who develop metastatic cancer would untimely surrender to this devastating disease due to the absence of effective treatment.1 Most often, breast tumors would consist of phenotypically diversified populations of breast cancer cells. During the treatment of breast cancer, steroid hormone receptors have been detected to be vital prognostic factors and predictive markers responding to endocrine therapy. Most breast cancers are hormone receptor-positive (HR+) tumors which account for around 70% of the total. Generally, HR+ breast tumors have a more favorable prognosis in contrast with HR-negative (HR–) ones. Among all, triple-negative breast cancer (TNBC) (estrogen receptor-negative, progesterone receptor-negative and HER2-negative) is especially perilous due to the lack of valid and specific therapy targeting these proteins involved.2–5
Small noncoding RNAs (ncRNAs) are short RNA molecules which would not be translated into proteins, including miRNAs and Piwi-interacting RNAs (piRNAs). In recent reports, a class of small ncRNAs that originate from tsRNA (tRNA-derived small RNAs) precursor sequences and mature sequences have drawn additional attention. In eukaryocyte, tRNAs are most likely transcribed through RNA polymerase III, typically 76–90 nucleotides in length. Pre-tRNAs and mature tRNAs would be extensively modified both before and after being exported to the cytoplasm for creating three different kinds of tRNA-derived ncRNAs, namely tRNA-derived small RNAs (tsRNAs), tRNA halves (tiRNAs) and tRNA-derived fragments (tRFs or tDRs).6,7 Most of the tsRNAs are produced in cell nucleus as a result of the pre-tRNA 3′ end cleavage. As validated by some early reports, after to be transported to the cytoplasm, tsRNAs would also work with both Ago and Piwi proteins to affect the expression of gene at a posttranscriptional level through 3′ UTR targeting, displaying a mechanism that is similar to miRNAs.8,9 By contrast, tiRNAs normally generated from angiogenin-mediated cleavage of mature cytoplasmic tRNAs which is generally triggered under stress conditions.10 Even though the biogenesis of TRFs still requires under further exploration, in the cytoplasm, a Dicer-dependent cleavage of mature tRNAs are widely suggested to be a feasible mechanism lead to the generation of TRF.6–9,11–13
Among cancer-related researches, extensive attention has been paid to the relationship between small ncRNAs and cancer development.14,15 As proven by an increasing number of reports, some certain small ncRNAs have played a critical part in the proliferation, metastasis and invasion of tumor cells. Apart from that, they have also been identified to participate in tumor development and vasculogenesis in several types of cancers.16–22 Particularly, the dysregulation of tsRNA in cancer has been increasingly studied in recent years.23,24 tsRNAs have also been demonstrated to be indispensable in the early and progressive stages of some certain types of cancers including B-cell lymphomas, chronic lymphocytic leukemia and lung carcinoma.9,24,25
However, the tsRNA expression profile in breast cancer has not been detected so far. Here, we used small RNA sequence to measure the expression level of small RNA in both breast cancer tissue and adjacent normal tissue. In this study, we noticed that tRNA-derived small RNAs (tsRNAs) were dysregulated in breast cancer. Especially, we observed that tsRNA-26576 was significantly upregulated (over 5-fold) in breast tumor compared with that of normal tissue. Moving on, we validated that tsRNA-26576 could promote breast cancer cell growth and inhibit cell apoptosis in vitro.
Materials and methods
Human tissue samples and cell lines
Tissues were acquired from patients who were conducting breast cancer surgery in Women’s Hospital School of Zhejiang University, immediately snap-frozen in liquid nitrogen, and stored at −80°C until RNA extraction. Prior patients’ consents and approval from the Institutional Research Ethics Committee were obtained. The breast cancer cell lines, namely MDA-MB-231, were acquired from the American Type Culture Collection. The cells were cultured in DMEM (MDA-MB-231) media (Gibco) added with 10% fetal bovine serum (Gibco) at 37°C with 5% CO2.
RNA extraction and sequencing
For mRNA sequencing, total RNA was extracted from the transfected MDA-MB-231 cells with TRizol reagent (Invitrogen) following the manufacturer instructions. Approximately 5 μg total RNA of each sample was used for the cDNA reverse transcription. After quantitative analysis and quality inspection, Illumina TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) was used to establish sequencing libraries. For small RNA sequencing, total RNA was extracted using the mirVana TM miRNA Isolation Kit (Ambion, Austin, TX) following the manufacturer’s instructions. For small RNA library establishment and deep sequencing, the 135–170 nt size range of RNA was enriched by polyacrylamide gel electrophoresis. RNA from each sample was subject to DNA sequencing with an Illumina HiSeq 2500 Sequencing System according to the manufacturer’s instructions.
Quantitative reverse transcription PCR
Real-time PCR was performed to determine the expression levels of candidate miRNAs applying the ABI 7300 Real-Time PCR System (Applied Biosystems, USA). RT-PCR was then carried out by using One Step TB Green™ PrimeScript™ PLUS RT-PCR Kit (Takara) following the manufacturer’s protocol. The reaction was conducted in triplicate with the 7500 real-time PCR system (Applied Biosystems). Meanwhile, the 2-ΔΔCt method was used for quantifying gene expression, and the expression of U6 was utilized as the internal control for tsRNA. The sequence information of each primer for RT-PCR that was used for gene expression analysis shown below:
U6-F 5’ CGATACAGAGAAGATTAGCATGGC 3’
U6-R 5’ AACGCTTCACGAATTTGCGT 3’”
tsrna-26576-FS 5’ CGTATTCGACGATCTCGAGAGG 3’
tsrna-26576-RS 5’ CGGAAACCGATCTAAAGAAACCT 3’
tsrna-21042-FS 5’ CAGCGACGATCGTCTAGTGGTTA 3’
tsrna-21042-RS 5’ GTCCGATCTACCCGGGCC 3’
tsrna-14773-FS 5’ ATCCGTATTCGACGATCGCAT 3’
tsrna-14773-RS 5’ TAACCGATCTTCCCGCGTG 3’
tsrna-10281-FS 5’ TATTCGACGATCGGCCGT 3’
tsrna-10281-RS 5’ CGTCCGATCTCAACGCAGA 3’
GAPDH -F 5’ AGAAGGCTGGGGCTCATT 3’
GAPDH -R 5’ TGCTAAGCAGTTGGTGGTG 3’
FAT4-F 5’ AATCCCTGGTTCTTTGGTAGCA 3’
FAT4-R 5’ TAATAGGATTCAGGAAAAAGATGCC 3’
SPEN-F 5’ GGATAGGTCCAGAAACAGAAAGTGA 3’
SPEN-R 5’ TGGAAGATGTTGCGAAGGTCA 3’
tsRNA mimics/inhibitor and transfections
Synthetic mimic or inhibitor of tsRNAs (GenePharma) were brought into cells through transient transfection with Lipo2000 (ThermoScientific), applying standard conditions and reaching a 50 nmol/L final concentration. In this study, a random sequence nontargeting mimic tsRNA (GenePharma) was introduced as a negative control. The sequence information of tsRNA-26576 or negative control mimic and inhibitor are shown as below:
tsRNA-26576 mimic: UCGAGAGGGGCUGUGCUCGCAAGGUUUCUUU
NC mimic: UUGUACUACACAAAAGUACUG
tsRNA-26576 inhibitor: AAAGAAACCUUGCGAGCACAGCCCCUCUCGA
inhibitor NC: CAGUACUUUUGUGUAGUACAA
Cell proliferation
Cell proliferation was evaluated with Cell Counting Kit-8 (CCK-8) (Beyotime Institute of Biotechnology). Transfection was performed as mentioned above. 48 hrs post-transfection, MDA-MB-231 cells were digested, resuspended and inoculated into 96-well plates. The plate was incubated for an appropriate length of time and then 10 μL of the CCK-8 solution was added to each well of the plate. The cells were then further incubated with CCK-8 for 1 hr at 37°C. The absorbancy was identified at 450 nm using microplate reader (Bio-Rad).
Transwell experiment
The cell migration assay was conducted using Corning Transwell chambers (corning 3422). In the invasion assay, the inserts coated with Matrigel were used to investigate cell invasive capability. Transfected MDA-MB-231 cells were washed with PBS and trypsinized with trypsin. The cells were then washed 2 times with serum-free medium and resuspended in serum-free medium, and 200 μL cell suspension (2×105 cells) of each group was supplemented to the upper chamber. 600 μL DMEM with 20% FBS was added to the well of the plate. The plate was then incubated in 5% CO2 at 37°C for 24 hrs. Then, all the cells in the upper chamber were removed, and the attached cells in the lower section were stained with 0.1% crystal violet.
Cell apoptosis assays
Transfection was performed as mentioned above. 48 hrs post transfection, MDA-MB-231 cells were digested, resuspended, centrifuged and washed with PBS. 195 µL cell suspension was taken and stained with 5 µL annexin V-FITC and 10 µL PI in the dark at room temperature for 30 mins, and measured with flow cytometry (FACSVerseTM, BD Biosciences).
Statistical analysis
Each experiment was repeated at least three times. Student’s t-test (two-tailed) was performed, and statistical significance level was set at α=0.05 (two-side).
Ethics approval and informed consent
This study was approved by the Human Ethics Committee of the Women’s hospital of Zhejiang University (Hangzhou, China). Prior patients’ consents and approval from the Institutional Research Ethics Committee were obtained. All patients gave written informed consent before participation in this study. All experiments were carried out in accordance with the principles of Declaration of Helsinki.
Result
Small RNA sequencing reveals a tsRNA expression profile in breast cancer
To identify the differential expressions of tsRNAs in breast tissue and noncancerous breast tissue, four paired samples from breast cancer patients (4 cancer tissue and four adjacent normal tissues) were profiled using small RNA sequencing. After filtering out the reads of both low-quality and masking-adaptor sequences, all the reads that are between 24 and 33 nucleotides in length were selected from 8 samples. For acquiring the different tsRNA expression levels, the small RNAs which match with known miRNA database (miRBase) and piRNA database (GtRNAdb) were removed. After that, with fold variation >2 and P-value <0.05 set as the threshold, a total of 263 tsRNAs were identified as significantly differentially expressed, of which 75 were upregulated and 188 were downregulated (
Functional annotation for target genes of differently expressed tsRNAs
For further exploring the biological function of these tsRNAs, the target genes of the differentially expressed tsRNA were predicted by using RNAhybrid; only the tsRNAs with max energy <−25 in RNAhybrid were included in this study. Furthermore, we carried out GO and KEGG pathway enrichment analyses for the tsRNA target genes for obtaining more reliable biological functions. GO analysis is frequently used to evaluate the enrichment of differently expressed genes (DEGs) regarding biological process (BP), cellular component (CC) and molecular function (MF). In this study, GO enrichment analysis was applied to investigate all DEGs between tumor tissues and normal tissues adjacent to these tumors. As revealed by the results, the most enriched GO term was related to the regulation of transcription, signal transduction and cell adhesion in the biological process analysis (Figure 2A). Besides, several signaling pathways including epidermal growth factor receptor signaling pathway, neurotrophin TRK receptor signaling pathway, insulin receptor signaling pathway and fibroblast growth factor receptor signaling pathway were affected as well (Figure 2B). On the other side, through the functional determination based on KEGG pathway database, the pathway in cancer was classified as the most significant pathway enriched by the targets of differently expressed tsRNAs, which implies that these differently expressed tsRNAs might act as a major factor functioning in the initiation and progression of breast cancer (Figure 2B).
tsRNA-26576 promotes cell growth in breast cancer cells
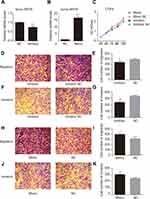
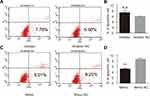
Among all the differently expressed tsRNAs between the cancer tissues and adjacent normal tissues, tsRNA-26576 displayed the most significant fold change. It was upregulated in the cancer tissues for about two-fold in our small RNA sequence data. We then detected its expression level in human breast tumor cell line: MDA-MB-231. The result showed that tsRNA-26576 was expressed in MDA-MB-231. This cell line was also used later in the following experiment to further determine the role of tsRNA-26576 in the progression of cancer. Synthetic, chemically modified mimic and inhibitor of tsRNA-26576 were introduced into MDA-MB-231 via transient transfection to overexpress and knockdown tsRNA-26576, respectively. For control, a non-targeting RNA (NC) was transfected as well. Both successful tsRNA overexpression and knockdown were detected by RT-PCR (Figure 3A and B). The cell growth rate of breast cancer cells transfected with tsRNA-26576 mimic and inhibitor were examined by CCK8 assay (Figure 3C). The CCK8 assay showed that tsRNA-26576 mimic would promote cell proliferation in MDA-MB-231 cells, while tsRNA-26576 inhibitor would lead to the suppression of cell growth (Figure 3C). These data suggested that tsRNA-26576 may be served as an indispensable factor in promoting breast cancer cell proliferation. Moreover, we tested the migration and invasion of cell through transwell migration and invasion assays, respectively, showing the suppression of tsRNA-26576 could remarkably inhibit the migration and invasion capability of MDA-MB-231 cells (Figure 3D–G), while the overexpression of tsRNA-26576 had an opposite effect (Figure 3H–K). The effects of tsRNA-26576 on cell apoptosis were assessed by flow cytometry analyses which indicate that the upregulation of tsRNA-26576 would lead to decreased cell apoptosis; however, tsRNA-26576 inhibitor did not influence cell apoptosis (Figure 4). This may be due to the relatively lower expression level of tsRNA-26576 in the MDA-MB-231 cells. So the inhibitor shows no remarkable effect. Altogether, these data suggest that tsRNA-26576 may play a crucial role in motivating cancer cell proliferation, migration, invasion and cell apoptosis inhibition.
Analysis of gene signatures of MDA-MB-231 cell responding to tsRNA-26576 inhibitor treatment
Understanding which genes are regulated by tsRNA-26576 is of great help for us to figure out the molecular mechanisms that how breast cancer could develop or progress. Based on our in vitro assays which identified the cell growth-promoting activities of tsRNA-26576 in breast cancer, we further used high-throughout sequencing technology to demonstrate how tsRNA-26576 regulates transcriptomic profile of breast cancer cell. We knock down tsRNA-26576 in MDA-MB-231 cells by delivering synthetic tsRNA-26576 inhibitor. After that, mRNA sequencing was applied to identify the differentially expressed genes. After delivering the tsRNA-26576, 483 genes were detected to be significantly altered by setting fold variation >2 and P-value <0.05 as the threshold. Among those, 154 genes were found to be down-regulated while 329 were upregulated in the inhibitor treatment group in comparing with control group (
Tumor suppressor gene SPEN and FAT4 are upregulated after knockdown of tsRNA-26576
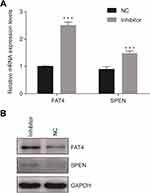
After suppression of tsRNA-26576 by delivering tsRNA-26576 inhibitor in MDA-MB-231 cells, we found a number of genes were upregulated by using mRNA sequencing. After overlapping the upregulated genes with tsRNA-26576 target genes, we enriched a number of genes; among these genes, SPEN and FAT4 were significantly upregulated. And based on the previous data, both of these two genes may be related to the breast cancer development, which are chosen for further study. In ERα-positive breast cancer cells, SPEN, as an ERα corepressor, has been determined to be a tumor suppressor protein.26,27 On the other side, FAT4,a cadherin-related protein, has been proven to play a great part in various biological processes. Previous data showed FAT4 could function as a tumor suppressor in TNBC and it may also be utilized as possible targets for the treatment of TNBC.28–30 In this study, SPEN and FAT4 were selected for RT-PCR and Western Blot to verify the species-specific gene expression levels which were quantified by RNA-Seq. As a result, SPEN and FAT4 were detected to be upregulated after the treatment of tsRNA-26576 inhibitor (Figure 6). These data showed that expression ratios from the RNA-seq were validated to be highly correlated with RT-PCR and Western Blot data.
Our data illustrate that tsRNA-26576 was upregulated in breast cancer indicating it might perform as a potential tumor initiator and enhance the tumor progression in human breast cancer. Therefore, tsRNA-26576 might also be served as a potential clinical predictive marker for breast cancer. Besides, after knockdown of tsRNA-26576, we found that several tumor suppressor genes were upregulated; based on that, these results might also propose a novel therapeutic solution for possible treatment of breast cancer through targeting tsRNA-26576.
Discussion
From the last decade, small RNAs have been increasingly explored in tumor-related research. They have been demonstrated to play a major part throughout the entire carcinogenic process including tumor onset, growth, metastasis and drug resistance development. Recently, accumulated evidence has validated that the dysregulation of tsRNAs could also affect various cellular processes which are associated with proliferation, metastasis and invasiveness of cancer cell. Those tsRNAs have also been proven to be related with tumor angiogenesis in some human malignant tumors, and more importantly, they have a direct impact on the performance of therapeutic interventions.16,18,31,32
Under stress conditions, tRFs have been demonstrated to mediate the stress-induced response and then influence several cellular reactions, such as cell proliferation through inactivating RNA.6,11–13,33 Previous data indicated that endogenous tRNA-derived fragments (tRFs) could suppress breast tumor progression through removing their 3′UTRs from the RNA-binding protein YBX1 and then suppress the stability of several oncogenic transcripts.33 However, the functions of tsRNAs in breast cancer remain have not yet been well understood. In this study, we, for the first time, report the tsRNA transcriptional profile of breast cancer. Also, we identified 263 tsRNAs that were differently expressed between breast cancer and normal tissue. These tsRNAs observably affect epidermal growth factor receptor signaling pathway, neurotrophin TRK receptor signaling pathway, insulin receptor signaling pathway and fibroblast growth factor receptor signaling pathway. All of them are important pathways for regulating cell growth and cancer development. Our data outline the significance of predictive and therapeutic values of cancer-related tsRNAs in breast cancer patients. Breast cancer has been classified as a type of heterogeneous disease with diversified morphological specificities and clinical features given genetic, epigenetic and transcriptomic variations.34,35 The different tsRNA expression levels in normal and tumor tissues may reflect the diversities associated with the transformed phenotypes or cell-type compositions amid the samples. For minimizing the impact from that, in this study we included both the samples from breast cancer patients and the samples of adjacent normal tissue (which are located around the cancer sample), together serving as paired samples.
Among there 263 tsRNAs, tsRNA-26576 was validated to be closely associated with breast cancer. In our study, tsRNA-26576 was detected to be significantly upregulated in breast cancer tissue. Moreover, we also found overexpress tsRNA-26576 could motivate cell proliferation and migration while inhibiting cell apoptosis. tsRNAs normally would remain stable and detectable in blood, and their dysregulations have been determined to be related with the development or progression of breast cancer, suggesting tsRNA-26576 may be an effective marker for breast cancer dia-gnosis.
Here, we found SPEN and FAT4 were upregulated after the treatment of tsRNA-26576 inhibitors, and both of them participate into a large number of biological processes as a tumor suppressor. Our data indicate that tsRNA-26576 could perform as an oncoprotein by inhibiting the expression of SPEN and FAT4. The RNA interference (RNAi) has been known as an effective mechanism to silence gene; it reveals a distinct capability in targeting cancer-related genes. Because of that, a great many of oncogene products related to tumorigenesis have been increasingly explored as possible targets in novel therapy based on RNAi strategy. In this study, our data also provide preclinical support for the use of small tsRNA-26576 RNA inhibitor therapies in breast cancer patients. Hopefully, the treatment of delivering tsRNA-26576 inhibitor might be utilized as one of the efficient therapy strategies for patients suffering from breast cancer.
Interestingly, the mRNA sequencing data showed that the most enriched pathways by GO analysis are related to nervous system development after tsRNA-26576 inhibitor treatment. Like miRNA, tsRNA expression also shows tissue-specific character; we speculated tsRNA-26576 may exist in the nervous system and regulate synapse assembly. In our mRNA sequencing data, we found several genes, which are critical for synapse function, were changed. For instance, NLGN3, SPOCK2, PCDHB9, PCLO, PCDHB11, BSN, ACHE were upregulated after tsRNA-26576a inhibitor treatment. All these genes may influence diverse steps of neurogenesis, and are important for the establishment and maintenance of specific neuronal connections, synaptic signal transmission and synaptic apoptosis. Further study may focus on tsRNA-26576 and nervous system development.
In conclusion, our study has illuminated the importance of tsRNAs in breast tumors and indicated that the dysregulation of these molecules during carcinogenesis might reveal a novel diagnostic tool as well as a potential therapy target for breast cancer.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
The work was supported by the National Science Foundation for Young Scientists of China (Grant No. 81502270).
Author contributions
All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74.
2. Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, Mcguire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177. doi:10.1126/science.3798106
3. Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;41(2):219–219.
4. Veer LJVT, Dai H, Vijver MJVD, et al. Gene expression profiling predicts clinical outcome of breast cancer. nature. 2002;415(6871):530.
5. Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2018;13(15 Pt 1):4429–4434.
6. Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23(22):2639–2649. doi:10.1101/gad.1837609
7. Venkatesh T, Suresh PS, Tsutsumi RJG. tRFs: miRNAs in disguise. Gene. 2016;579(2):133–138.
8. Canella D, Praz V, Reina JH, Cousin P, Hernandez NJGR. Defining the RNA polymerase III transcriptome: genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20(6):710.
9. Haussecker D, Huang YA, Parameswaran P, Fire AZ, Kay MAJR. Human tRNA-derived small RNAs in the global regulation of RNA silencing. Rna. 2010;16(4):673–695. doi:10.1261/rna.2000810
10. Fu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583(2):437–442. doi:10.1016/j.febslet.2008.12.043
11. Cole C, Sobala A, Lu C, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. Rna. 2009;15(12):2147–2160. doi:10.1261/rna.1738409
12. Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138(2):215–219. doi:10.1016/j.cell.2009.07.001
13. Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10(12):1798–1806. doi:10.4161/rna.27177
14. Bahrami A, Aledavood A, Anvari K, et al. The prognostic and therapeutic application of microRNAs in breast cancer: tissue and circulating microRNAs. J Cell Physiol. 2017;233:2.
15. Nassar FJ, Nasr R, Talhouk RJPT. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Therapeut. 2017;172:34–49. doi:10.1016/j.pharmthera.2016.11.012
16. Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer. 2005;65(16):7065.
17. Zhang L, Huang J, Yang N, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103(24):9136–9141. doi:10.1073/pnas.0508889103
18. *
19. Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Res. 2007;8(10):1–16.
20. Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci. 2006;103:2257–2261. doi:10.1073/pnas.0510565103
21. Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi:10.1016/j.ccr.2006.01.025
22. Yu F, Yao H, Zhu P, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi:10.1016/j.cell.2007.10.054
23. Balatti V, Pekarsky Y, Croce CM. Role of the tRNA-derived small RNAs in cancer: new potential biomarkers and target for therapy. Adv Cancer Res. 2017;135:173.
24. Balatti V, Nigita G, Veneziano D, et al. tsRNA signatures in cancer. Proc Natl Acad Sci. 2017;114(30):201706908.
25. Maute RL, Christof S, Pavel S, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci. 2013;110(4):1404–1409.
26. Légaré S, Cavallone L, Mamo A, et al. The Estrogen receptor cofactor SPEN functions as a tumor suppressor and candidate biomarker of drug responsiveness in hormone-dependent breast cancers. Cancer Res. 2015;75(20):4351–4363. doi:10.1158/0008-5472.CAN-14-3475
27. Légaré S, Chabot C, Basik M. SPEN, a new player in primary cilia formation and cell migration in breast cancer. Breast Cancer Res. 2017;19(1):104.
28. Hou L, Chen M, Zhao X, et al. FAT4 functions as a tumor suppressor in triple-negative breast cancer. Tumour Biol 2016;1–7.
29. Katoh M. Function and cancer genomics of FAT family genes (review). Int J Oncol. 2012;41(6):1913–1918. doi:10.3892/ijo.2012.1669
30. Katoh Y, Katoh M. Comparative integromics on FAT1, FAT2, FAT3 and FAT4. Int J Mol Med. 2006;18(3):523–528.
31. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi:10.1038/nrc1997
32. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi:10.1038/nrg2843
33. Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161(4):790–802. doi:10.1016/j.cell.2015.02.053
34. Müller V, Stahmann N, Riethdorf S, et al. Circulating tumor cells in breast cancer: correlation to bone marrow micrometastases, heterogeneous response to systemic therapy and low proliferative activity. Clin Cancer Res. 2005;16(4):331–332.
35. Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13(11):674.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.