Back to Journals » OncoTargets and Therapy » Volume 14
Small Proline-Rich Protein 2B Facilitates Gastric Adenocarcinoma Proliferation via MDM2-p53/p21 Signaling Pathway
Authors Yao L, Yan J, Cheng F, Gan L, Huang Y, Zheng L, Fang N
Received 10 September 2020
Accepted for publication 13 January 2021
Published 26 February 2021 Volume 2021:14 Pages 1453—1463
DOI https://doi.org/10.2147/OTT.S281032
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Takuya Aoki
Ling Yao,1 Jinhua Yan,2 Fei Cheng,1 Lihong Gan,1 Yaqin Huang,1 Li Zheng,1 Nian Fang1
1Department of Gastroenterology, Third Affiliated Hospital of Nanchang University, Nanchang, 330008, Jiangxi Province, People’s Republic of China; 2Department of Hematology, Third Affiliated Hospital of Nanchang University, Nanchang, 330008, Jiangxi Province, People’s Republic of China
Correspondence: Nian Fang
Department of Gastroenterology, Third Affiliated Hospital of Nanchang University, No. 128 Xiangshan North Road, Nanchang, 330008, Jiangxi Province, People’s Republic of China
Tel +86-0791-88862387
Email [email protected]
Background: The small proline-rich protein 2B (SPRR2B) was firstly reported as a member of the cross-linked envelope protein in keratinocytes. The effect of SPRR2B in gastric adenocarcinoma (GC) remains unclear. This study initially explored the clinical significance of SPRR2B in GC patients as well as its role in tumor progression.
Methods: Immunohistochemistry was performed to characterize the expression of SPRR2B in GC tissues and adjacent tissues. The relationship between SPRR2B expression and clinicopathological features of GC patients was analyzed by Chi-square test. Kaplan-Meier method and Cox regression analyses were utilized to identify the prognostic factors of GC. Overexpression and knockdown assays were conducted to investigate possible signaling pathways downstream of SPRR2B. Flow cytometry assays were performed to evaluate cell cycle and apoptosis. Xenograft experiments were performed to validate tumor-related role of SPRR2B in vivo.
Results: Both mRNA and protein levels of SPRR2B in cancerous tissue were significantly higher than those in non-cancerous tissues. Meanwhile, SPRR2B expression was significantly associated with tumor size and tumor stage. Survival analysis revealed SPRR2B as one of the independent prognosis factors for overall survival of GC patients. Cellular and xenografts data implicated that silencing SPRR2B blocked the cell cycle of GC cells perhaps through MDM2-p53/p21-CDK1 pathway, while overexpressing SPRR2B exhibited opposite effects.
Conclusion: Our data suggest that SPRR2B may serve as a novel prognostic marker in GC, which functions at least partially by MDM2-p53/p21-CDK1 signaling pathway.
Keywords: gastric adenocarcinoma, MDM2, prognosis, proliferation, SPRR2B
Introduction
Gastric adenocarcinoma (GC) accounts for about 90% of stomach cancers and is characterized with poor prognosis. Current treatment strategies for GC patients include surgical resection, adjuvant chemotherapy, neoadjuvant chemotherapy, radiation therapy, and targeted therapy.1 Despite great achievements in GC treatment have been obtained during the past decades, the five-year overall survival for patients with advanced gastric cancer are far from satisfied (ranging from 8% to 20%).2 Therefore, it is still an urgent demand to further investigate GC progression mechanisms, identify effective prognostic biomarkers, as well as develop more alternative therapies.
The small proline rich protein (SPRR) gene family encodes a conserved group of cornified envelope (CE) proteins that are part of the human epidermal differentiation complex (EDC).3 The formation of the cornified envelope during the late stages of epidermal differentiation is essential for epidermal barrier function and protects the body against environmental attack and water loss.4 There are three subclasses of SPRR gene family, including the SPRR1 (contains SPRR1A and SPRR1B),5 SPPR2 (contains SPRR2A-2K), and SPRR3.6 Although SPRRs were initially identified to play roles in keratocyte differentiation, its function in other cells has also been reported.7 For example, SPRR1 participates in tobacco smoke-induced squamous metaplasia in rat nasal epithelia.8 SPRR1 is also recognized as a stress-inducible cardioprotective protein.9 Besides, SPRR2 was reported to be an allergen- and IL-13-induced protein that is involved in gastrointestinal and bronchial inflammations.10 Both SPRR1 and SPRR2A are increased in the mucosal from patients with chronic rhinosinusitis, which is perhaps downstream of TNF alpha modulation and lead to epithelial barrier dysfunction.11
Of note, SPRR proteins also participate in carcinogenesis and cancer progression. For example, SPRR2 is differentially affected by carcinogenic transformation in various squamous cell carcinomas.12 However, SPRR proteins seemed to play distinct roles in different cancer types. On one hand, SPRR1B and SPRR2A were reported to impair distant metastasis of tone squamous cell carcinomas cells, thus acting as anti-tumor proteins.13 On the other hand, expression levels of SPRR1, SPRR2E and SPRR3 were increased in lung squamous carcinoma, indicating their positive associations with lymph node metastasis.14 Similarly, SPRR3 promotes tumor cell proliferation in less advanced stages of breast cancer, which is associate with HER2/neu status.15 However, our knowledge about the expression and role of SPRRs in gastric cancer is limited.
Here we initially reported that SPRR2B was highly expressed and was an independent predictor of poor prognosis in gastric adenocarcinoma. We also revealed that SPRR2B can facilitate the cell cycle of GC cells and enhance xenograft growth, thus highlighting the potential significance of targeting SPRR2B in cancer treatment.
Materials and Methods
Patients and Samples
This study was conducted ethically in accordance with the World Medical Association Declaration of Helsinki and was approved by the Third Affiliated Hospital of Nanchang University Ethic Committee. Here we enrolled two independent retrospective cohorts. The first cohort was only used for RT-qPCR analyses and contained 32 paired fresh-frozen GC samples and adjacent noncancerous samples. The second cohort was used for IHC detection and survival analyses, comprising 128 formalin-fixed paraffin-embedded (FFPE) GC samples from patients whose follow-up data were intact. All diagnoses were based on histopathological tests after tumor resection. All participants in the two cohorts signed the written informed consent documents.
RNA Extraction and RT-qPCR
We firstly extracted mRNA from fresh-frozen tissues following the standard manufacturer's procedure. Then, isolated mRNA was used for reverse transcription using a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific, USA). After reverse transcription, the cDNA products were used for RT-qPCR assay using the SYBR Green PCR Master Mix (Thermo Fisher Scientific, USA) according to the manufacturer’s procedure.16 We choose GAPDH as endogenous reference gene, and qPCR primers were as follows:
SPRR2B-Forward: 5′‐CCCACCCTGCCAGCCAAAGTA‐3′
SPRR2B-Reverse: 5′‐CATGCCCAGGTGAAAGACAGACA‐3′17
GAPDH-Forward: 5′- GTCTCCTCTGACTTCAACAGCG-3′
GAPDH-Reverse: 5′- ACCACCCTGTTGCTGTAGCCAA −3′
Immunohistochemistry (IHC) Staining
IHC staining was performed to evaluate SPRR2B protein level in human tissues following the manufacturer instructions. Sample slides were sectioned at 8 μm. Slide sections were next dried and deparaffinized, then taken for antigen retrieval within citrate buffer. Then, slide sections were blocked and incubated with the primary SPRR2B antibody at 4°C overnight. Finally, the slide sections were washed and incubated with secondary antibody and DAB substrates were utilized to detect immunoreactivities.18 Negative control was conducted by using PBS instead of primary antibody.
Evaluation of IHC Staining
The IHC staining results were scored by two pathologists independently. IHC staining intensity was scored as: 1 (negative), 2 (weak staining, slight yellow), 3 (moderate staining, dark yellow), and 4 (strong staining, dark brown). The proportion of positive cells was scored as: 1 (0–25%); 2 (25–50%); 3 (50–75%); 4 (75–100%). Finally, the total score of IHC staining was calculated by multiplying these two scores above (ranging 1–16).
Cell Culture and Transfection
Human gastric epithelium GES-1 cell line was purchased from the Shanghai Cell Bank of the Chinese Academy of Science (Shanghai, China) and kept in DMEM medium. AGS cell line was obtained from ATCC and kept in F-12K medium. MKN28 and MKN45 cells were obtained from ATCC and maintained in RPMI 1640 medium. All medium was supplemented with 10% fetal bovine serum and penicillin-streptomycin antibiotics.
Human SPRR2B-shRNA (sc-88,442-SH) and control-shRNA (sc-108,060) plasmids were obtained from Santa Cruz Biotechnology. MKN45 cells were transfected with shRNA plasmids using shRNA Plasmid Transfection Reagent (sc-108,061, Santa Cruz).19 Transfected cells were selected by using puromycin. Specific siRNAs targeting MDM2 (sc-29,394) and SPRR2B (sc-88,442) were used for transient transfection, using non-specific scramble siRNA (sc-37,007) as negative control. For overexpression assays, pcDNA3.1-vector, pcDNA3.1-SPRR2B, and pcDNA3.1-SPRR2B-optimized plasmids were synthesized by Gene Pharma (Shanghai, China). The pcDNA3.1-SPRR2B-optimized (pcDNA-SPRR2B°pt) construct was generated by mutating the SPRR2B-siRNA targeted region of wild type SPRR2B and was used for rescue experiments.
Protein Extraction and Western Blot
Transfected or non-transfected cells were cultured for 48–72 hours and then harvested with cold NP-40 lysis buffer containing protease inhibitors. Lysates were collected and then centrifuged at 12,000 g for 20 min to obtain the supernatants. After quantification using a BCA kit (Pierce, Pittsburgh, PA), proteins were denatured and resolved by SDS-PAGE electrophoresis and then transferred onto the NC membrane for immunoblotting. The NC membranes were blocked with 5% nonfat milk and then incubated with primary antibodies at 4°C overnight. Finally, horseradish peroxidase-conjugated secondary antibodies were added to incubate for another 1 h at room temperature followed by signal detection using ECL substrate solution and X-ray films.20
Cell Apoptosis and Cell Cycle Assays
The apoptosis assay was conducted as we previously described.21 Briefly, transfected cells were trypsinized and collected by centrifugation, then resuspended in 500 µL of buffer and incubated with Annexin V-FITC/PI kit for 15 min. The apoptotic rate was detected using a flow cytometer (FACSCalibur, BD Biosciences). Each experiment was performed for three times.
The flow cytometry strategy was also used to test cell cycle as previously described.22 Briefly, cells were trypsinized and collected by centrifugation, and then cells were incubated with propidium iodide (PI) stain buffer for 10 min and then treated with 200 µL of PBS, 200 µL of RNase A and 200 µL of PI for another 10 min at 4°C in dark. Proportions of cells in each of the cell cycle phases were analyzed using the flow cytometer. Each experiment was performed three times.
Xenografts Assay
The xenograft animal experiments were approved by the Ethics Committee of our hospital and performed in accordance with the Guide for the Care and Use of Laboratory Animals (8th edition). 5×105 MKN45 stably transfected cells were subcutaneously injected into the flank of the male BALB/c nude mice at 4–5 weeks old (approximately 20–22g, randomly grouped).23 The tumor size in mice was measured by a vernier caliper every 5 days for 3 weeks, and the tumor volume was calculated by the following formula: (π x length x width2)/6. At 21 days after injection, all mice were sacrificed, the tumor tissues were isolated and weighed.
Statistical Analyses
Statistical analyses were performed using the SPSS 22.0 Software. The associations between SPRR2B protein level and clinical characteristics were evaluated through Chi-square test. Kaplan–Meier analysis and Log rank test were used to plot and analyze the overall survival curves of enrolled GC patients. Independent prognostic factors were identified by using multivariate Cox regression model. Student’s t-test and One-way ANOVA test were conducted to compare the differences between groups for in vitro and in vivo experiments. P<0.05 was considered statistically significant.
Results
Patients’ Characteristics
Among the 32 cases in the first cohort, which was only used for RT-qPCR analysis, there were: 19 males and 13 females; 16 T1-T2 stages and 16 T3-T4 stages; 5 TNM stage I, 16 TNM stage II, and 11 TNM stage III.
Among the 128 cases in the second cohort, which was used for IHC test and survival analysis, there were 46 females and 82 males (Table 1). Tumors were located in the upper 1/3 gastric for 26 cases, middle 1/3 for 47 cases, and lower 1/3 for the other 55 cases. Most tumor size was smaller than or equal to 5.0 cm in diameter (79 cases), while the other 49 cases with tumor size larger than 5.0 cm. As for histological grade, poor differentiation tumors were characterized for 61 cases, moderate differentiation for 51 cases, and only 15 cases with well differentiation grade. Besides, 76 tumors were classified with T1-T2 infiltration stage, while 52 cases with T3-T4 stage according to the AJCC cancer staging system (8th edition). Consistently, 80 cases were pathologically staged with TNM stage I–II, and 48 cases with TNM stage III. The median follow-up time was 42 months (ranging 5–79 months) for these 128 cases, with a 5-year overall survival rate at 45.4%.
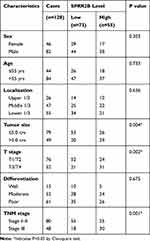 |
Table 1 Correlations Between SPRR2B Expression with Patients’ Characteristics |
SPRR2B is Upregulated in Gastric Adenocarcinoma on Both mRNA and Protein Levels
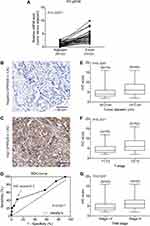
To explore the expression pattern of SPRR2B in GC, we initially tested the mRNA levels of SPRR2B in 32 pairs of fresh GC samples together with their adjacent gastric tissues using RT-qPCR method. As a result, SPRR2B-mRNA level was substantially higher in GC compared with adjacent noncancerous gastric tissues (Figure 1A, 3.93 folds, P<0.001).
Moreover, we tested the SPRR2B protein level in another 128 GC samples by IHC immunostaining, which showed its predominantly cytosol and membrane localization (Figure 1B and C). Consistent with RT-qPCR results, IHC data demonstrated that the protein level of SPRR2B was significantly up-regulated in some GC samples compared with adjacent noncancerous samples. Therefore, we sub-grouped these 128 patients into high-SPRR2B group (IHC score ≥5) and low-SPRR2B group (IHC score <5) according to the receiver operating characteristic (ROC) curve (Figure 1D, P<0.001).
SPRR2B is Associated with Clinical Characteristics of Gastric Adenocarcinoma Patients
Chi-square test was next conducted to assess the clinical significance of SPRR2B in GC (Table 1). Accordingly, patients with larger tumor size (Figure 1E, P=0.008), advanced T stage (Figure 1F, P<0.001), or advanced TNM stage (Figure 1G, P=0.025) exhibited higher SPRR2B protein levels. In contrast, neither tumor localization nor tumor differentiation showed significant association with SPRR2B level (both P>0.05).
High SPRR2B Predicts Unfavorable Overall Survival of GC Patients
To further investigate the prognostic role of SPRR2B in GC patients, we performed Kaplan–Meier analysis together with Log rank test (Figure 2, Table S1). Of note, GC patients with higher SPRR2B level showed poorer overall survival (mean 38.7 ± 2.7 months) compared to those with lower or negative SPRR2B expression (mean 58.6 ± 2.7 months, Figure 2A). Besides, tumor size was identified as another unfavorable prognosis factor (P=0.033). As expected, either advanced T stage or TNM stage was also negatively correlated with patients’ overall survival time (P=0.026 and P<0.001, respectively).
We then subjected these significant risk factors above into a Cox regression model for multivariate analysis (Table 2). As a result, TNM stage was identified as an independent unfavorable prognostic factor in GC patients (HR=2.419, 95% CI=1.118–5.233, P=0.025). Moreover, our data revealed SPRR2B as a novel independent prognostic biomarker in GC for the first time (HR=2.246, 95% CI=1.192–4.230, P=0.012), further highlighting its clinical significance.
 |
Table 2 Multivariate Analysis |
SPRR2B Promotes Cell Cycle by MDM2 Downstream Signaling Pathways
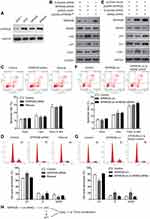
By comparing the expression level of SPRR2B in nontumorous GES-1 cells and several gastric adenocarcinoma cell lines (AGS, MKN28, MKN45), we found that SPRR2B protein was upregulated in gastric cancer cells (Figure 3A), which was consistent with immunohistochemistry results of clinical specimens. We were also interested to further explore possible signaling pathways downstream of SPRR2B in modulating malignancy progression. Literature research reminded us that SPRR2B had crosstalk with MDM2/p53 pathway in fibroblast proliferation during heart failure.24 Considering the involvement of MDM2/p53 in tumor development, we next focused on this signaling pathway. Silencing SPRR2B can significantly downregulate MDM2 and CDK1 levels while upregulate p53 and p21 protein levels in MKN45 cells (Figure 3B). However, these alterations can be rescued by re-expressing the modified SPRR2B plasmids (SPRR2B°pt). Transfected cells were then subjected to apoptosis test (Figure 3C). Accordingly, neither the early apoptosis nor late apoptosis process was significantly affected by SPRR2B-knockdown. Nevertheless, cell cycle tests showed a blockage of cell cycle in SPRR2B-siRNA transfected cells, which can be rescued by overexpressing SPRR2B°pt (Figure 3D). The changes in cell cycle were consistent with CDK1 level alterations in Figure 2B.
In addition, we overexpressed wild type SPRR2B and showed its positive effect on upregulating CDK1 level (Figure 3E). Simultaneously silencing MDM2 can abolish the alterations of p53, p21, and CDK1 proteins, indicating SPRR2B may function in a MDM2-dependent manner. On one hand, overexpressing SPRR2B showed no significant effect on apoptosis process (Figure 3F). On the other hand, overexpressing SPRR2B facilitated MKN45 cells enter the G2/M phase of cell cycle, while silencing MDM2 abolished this effect (Figure 3G). Therefore, we hypothesized that SPRR2B may promote tumor proliferation at least partially through MDM2-p53/p21-CDK1 signaling pathway (Figure 3G).
SPRR2B Facilitates Gastric Adenocarcinoma Growth in vivo
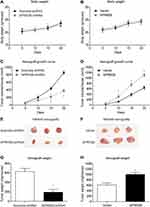
To further explore the effect of SPRR2B on tumor growth in vivo, xenograft nude mice model was established using stably transfected MKN45 cells. Mice were randomly divided into four groups and injected with transfected cells, and then the weight of each mouse was monitored every five days (Figure 4A and B). The xenograft growth curves showed that knockdown of SPRR2B led to a significant decrease in tumor proliferation compared to the scramble-shRNA group (Figure 4C). In contrast, overexpressing SPRR2B significantly enhanced the growth of xenografts (Figure 4D). Consistent with growth curves, the weight of xenografts was also smaller in SPRR2B-shRNA group than that of control group, while SPRR2B-overexpression exhibited an opposite effect (Figure 4E–H). Taken together, our results indicated that SPRR2B can positively regulate tumor growth in vivo.
Discussion
Here we firstly showed that SPRR2B is highly expressed in gastric adenocarcinoma from both mRNA and protein levels. According to our data, higher SPRR2B is positively correlated with larger tumor size and advanced TNM stage, suggesting its role in tumor progression. The participate of SPRR2B in gastric cancer is consistent with previous studies on that other SPRRs may also modulate malignancy progression, such as SPRR1A,25–28 SPRR1B,29–31 SPRR2A,32–35 and SPRR3.36–39
However, SPRRs can serve as either tumor suppressors39 or oncoproteins38 in different malignancies. Therefore, we suggest that SPRRs may play completely different functions in distinct cell types. Here we focused on gastric adenocarcinoma cells and demonstrated that SPRR2B can enhance cell proliferation capacity. According to the immunoblotting data, silencing SPRR2B can downregulate the MDM2 protein, a E3 ligase upstream of p53 tumor suppression, subsequently impair p53 and p21 degradation. Considering the critical role of p53/p21 in cell proliferation, we proposed that SPRR2B may promote gastric cancer proliferation at least partially through MDM2-p53/p21 signaling pathway, which was later validated by rescue experiments. Our data is consistent with its role in driving stress-dependent p53 degradation and fibroblast proliferation in heart failure.24 Similarly, overexpressing SPRR3 led to increased MDM2 activity and decreased the p53 protein level in breast cancer cells, which is downstream of AKT and MAPK pathways.15 Intriguingly, SPRR2A was reported to enhance p53 deacetylation and downregulates p21 promoter activity in epithelial cells,40 which is completely opposite to the role of SPRR2B in gastric adenocarcinoma or heart fibroblast. Therefore, the multifaced roles of SPRRs in biological processes need further systematic illumination.
There are several limitations in our study that can be further improved in following studies. Firstly, all clinical specimens were collected from a single medical center and thus may lead to regional bias. A multi-center cooperation would hopefully help validate our major conclusions. Secondly, the more detailed mechanism of how SPRR2B promote gastric cancer proliferation is not fully demonstrated. We suggest that cell cycle is involved according to flow cytometry results and the consistent alteration of checkpoint protein CDK1. This study is mainly focused on revealing clinical significance of SPRR2B in gastric cancer, further studies would be necessary to systematically investigate the underlying molecular signaling pathways to provide more evidence for novel therapy development.
Conclusion
Our results demonstrated that the expression level of SPRR2B was elevated in gastric adenocarcinoma tissues and significantly correlated with poor overall survival of gastric cancer patients. SPRR2B promotes gastric cancer cell proliferation both in vitro and in vivo, which may at least partially through MDM2-p53/p21 signaling pathway.
Data Sharing Statement
Data will be available upon request from Dr. Nian Fang.
Funding
This study was supported by National Natural Science Foundation of China (No. 81660408).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet. 2020;396(10251):635–648. doi:10.1016/S0140-6736(20)31288-5
2. Chen Y, Tian P, Liu Y. P53 and protein phosphorylation regulate the oncogenic role of epithelial cell transforming 2 (ECT2). Med Sci Monit. 2017;23:3154–3160. doi:10.12659/MSM.905388
3. Steven AC, Steinert PM. Protein composition of cornified cell envelopes of epidermal keratinocytes. J Cell Sci. 1994;107(Pt 2):693–700.
4. Cabral A, Voskamp P, Cleton-Jansen A-M, et al. Structural organization and regulation of the small proline-rich family of cornified envelope precursors suggest a role in adaptive barrier function. J Biol Chem. 2001;276(22):19231–19237. doi:10.1074/jbc.M100336200
5. Reddy SPM, Konkin T, Wu R. Structure and organization of the genes encoding mouse small proline-rich proteins, mSPRR1A and 1B. Gene. 1998;224(1–2):59–66. doi:10.1016/S0378-1119(98)00507-1
6. Fischer DF, Sark MWJ, Lehtola MM, et al. Structure and evolution of the human SPRR3 gene: implications for function and regulation. Genomics. 1999;55(1):88–99. doi:10.1006/geno.1998.5622
7. Carregaro F, Stefanini ACB, Henrique T, et al. Study of small proline-rich proteins (SPRRs) in health and disease: a review of the literature. Arch Dermatol Res. 2013;305(10):
8. Tesfaigzi J, Th’ng J, Hotchkiss JA, et al. A small proline-rich protein, SPRR1, is upregulated early during tobacco smoke-induced squamous metaplasia in rat nasal epithelia. Am J Respir Cell Mol Biol. 1996;14(5):478–486. doi:10.1165/ajrcmb.14.5.8624253
9. Pradervand S, Yasukawa H, Muller OG, et al. Small proline-rich protein 1A is a gp130 pathway- and stress-inducible cardioprotective protein. EMBO J. 2004;23(22):4517–4525. doi:10.1038/sj.emboj.7600454
10. Zimmermann N, Doepker MP, Witte DP, et al. Expression and regulation of small proline-rich protein 2 in allergic inflammation. Am J Respir Cell Mol Biol. 2005;32(5):428–435. doi:10.1165/rcmb.2004-0269OC
11. Ramakrishnan VR, Gonzalez JR, Cooper SE, et al. RNA sequencing and pathway analysis identify tumor necrosis factor alpha driven small proline-rich protein dysregulation in chronic rhinosinusitis. Am J Rhinol Allergy. 2017;31(5):283–288. doi:10.2500/ajra.2017.31.4457
12. Lohman FP, Medema JK, Gibbs S, et al. Expression of the SPRR cornification genes is differentially affected by carcinogenic transformation. Exp Cell Res. 1997;231(1):141–148. doi:10.1006/excr.1996.3458
13. Fang Z. Long non-coding RNA MALAT-1 modulates metastatic potential of tongue squamous cell carcinomas partially through the regulation of small proline rich proteins. BMC Cancer. 2016;16(1):706.
14. Xiong Y, Li M, Zhang P, et al. Study on genetype in lung squamous carcinoma by high-throughput of transcriptome sequence. Zhongguo Fei Ai Za Zhi. 2017;20(11):727–731. doi:10.3779/j.issn.1009-3419.2017.11.01
15. Kim JC, Yu JH, Cho YK, et al. Expression of SPRR3 is associated with tumor cell proliferation in less advanced stages of breast cancer. Breast Cancer Res Treat. 2012;133(3):909–916. doi:10.1007/s10549-011-1868-5
16. Tan W, Pan M, Liu H, et al. Ergosterol peroxide inhibits ovarian cancer cell growth through multiple pathways. Onco Targets Ther. 2017;10(p):3467–3474. doi:10.2147/OTT.S139009
17. Takei K, Mitoma C, Hashimoto‐Hachiya A, et al. Antioxidant soybean targlyteer rescues T-helper-mediated downregulation of filaggrin expression via aryl hydrocarbon receptor. J Dermatol. 2015;42(2):171–180. doi:10.1111/1346-8138.12717
18. Hou S, Du P, Wang P, et al. Significance of MNK1 in prognostic prediction and chemotherapy development of epithelial ovarian cancer. Clin Transl Oncol. 2017;19(9):1107–1116. doi:10.1007/s12094-017-1646-x
19. Zhang Q, Fan H, Liu H, et al. WNT5B exerts oncogenic effects and is negatively regulated by miR-5587-3p in lung adenocarcinoma progression. Oncogene. 2020;39(7):1484–1497. doi:10.1038/s41388-019-1071-4
20. Xu YF. Sprouty2 suppresses progression and correlates to favourable prognosis of intrahepatic cholangiocarcinoma via antagonizing FGFR2 signalling. J Cell Mol Med. 2018;22(11):5596–5606.
21. Jian J, Li S, Liu L, et al. XPD inhibits cell growth and invasion and enhances chemosensitivity in esophageal squamous cell carcinoma by regulating the PI3K/AKT signaling pathway. Int J Mol Med. 2020;46(1):201–210. doi:10.3892/ijmm.2020.4593
22. Jian J, Li S, Fang N, et al. Pim-3 alleviates lipopolysaccharide-stimulated AR42J pancreatic acinar cell injury via improving the inflammatory microenvironment. Exp Ther Med. 2019;18(6):4427–4435. doi:10.3892/etm.2019.8094
23. Sun R, Liu Z, Qiu B, et al. Annexin10 promotes extrahepatic cholangiocarcinoma metastasis by facilitating EMT via PLA2G4A/PGE2/STAT3 pathway. EBioMedicine. 2019;47:142–155. doi:10.1016/j.ebiom.2019.08.062
24. Burke RM, Lighthouse JK, Quijada P, et al. Small proline-rich protein 2B drives stress-dependent p53 degradation and fibroblast proliferation in heart failure. Proc Natl Acad Sci U S A. 2018;115(15):E3436–e3445. doi:10.1073/pnas.1717423115
25. Chen G, Li G, Luo M, et al. Clinical significance of SPRR1A expression in progesterone receptor-positive breast cancer. Tumour Biol. 2015;36(4):2601–2605. doi:10.1007/s13277-014-2879-8
26. Pavón MA, Arroyo-Solera I, León X. The combined use of EFS, GPX2, and SPRR1A expression could distinguish favorable from poor clinical outcome among epithelial-like head and neck carcinoma subtypes. Head Neck. 2019;41(6):1830–1845.
27. Deng Y, Zheng X, Zhang Y, et al. High SPRR1A expression is associated with poor survival in patients with colon cancer. Oncol Lett. 2020;19(5):3417–3424. doi:10.3892/ol.2020.11453
28. Zhang H, Gao J, Zhao Z, et al. Clinical implications of SPRR1A expression in diffuse large B-cell lymphomas: a prospective, observational study. BMC Cancer. 2014;14(1):333. doi:10.1186/1471-2407-14-333
29. Patterson T, Vuong H, Liaw Y-S, et al. Mechanism of repression of squamous differentiation marker, SPRR1B, in malignant bronchial epithelial cells: role of critical TRE-sites and its transacting factors. Oncogene. 2001;20(5):634–644. doi:10.1038/sj.onc.1204134
30. Michifuri Y, Hirohashi Y, Torigoe T, et al. Small proline-rich protein-1B is overexpressed in human oral squamous cell cancer stem-like cells and is related to their growth through activation of MAP kinase signal. Biochem Biophys Res Commun. 2013;439(1):96–102. doi:10.1016/j.bbrc.2013.08.021
31. Oyesanya RA, Bhatia S, Menezes ME, et al. MDA-9/Syntenin regulates differentiation and angiogenesis programs in head and neck squamous cell carcinoma. Oncoscience. 2014;1(11):725–737. doi:10.18632/oncoscience.99
32. Zucchini C, Strippoli P, Rosati G, et al. Expression analysis and mutational screening of the epithelium-specific ets gene-1 (ESE-1) in patients with squamous anal cancer. Int J Oncol. 2000;17(2):265–270. doi:10.3892/ijo.17.2.265
33. Specht S, Isse K, Nozaki I, et al. SPRR2A expression in cholangiocarcinoma increases local tumor invasiveness but prevents metastasis. Clin Exp Metastasis. 2013;30(7):877–890. doi:10.1007/s10585-013-9589-2
34. Mizuguchi Y, Isse K, Specht S, et al. Small proline rich protein 2a in benign and malignant liver disease. Hepatology. 2014;59(3):1130–1143. doi:10.1002/hep.26889
35. Nisa L, Barras D, Medová M. Comprehensive genomic profiling of patient-matched head and neck cancer cells: a preclinical pipeline for metastatic and recurrent disease. Mol Cancer Res. 2018;16(12):1912–1926. doi:10.1158/1541-7786.MCR-18-0056
36. Li Q, Wang Y, Hu R. Dysregulation of SPRR3/Mir-876-3p axis contributes to tumorigenesis in non-small-cell lung cancer. Onco Targets Ther. 2020;13:2411–2419.
37. de a Simão T, Souza-Santos PT, de Oliveira DSL. Quantitative evaluation of SPRR3 expression in esophageal squamous cell carcinoma by qPCR and its potential use as a biomarker. Exp Mol Pathol. 2011;91(2):584–589. doi:10.1016/j.yexmp.2011.06.006
38. Cho DH, Jo YK, Roh SA, et al. Upregulation of SPRR3 promotes colorectal tumorigenesis. Mol Med. 2010;16(7–8):271–277. doi:10.2119/molmed.2009.00187
39. Chen BS. Decreased expression of SPRR3 in Chinese human oesophageal cancer. Carcinogenesis. 2000;21(12):2147–2150. doi:10.1093/carcin/21.12.2147
40. Mizuguchi Y, Specht S, Lunz JG, et al. SPRR2A enhances p53 deacetylation through HDAC1 and down regulates p21 promoter activity. BMC Mol Biol. 2012;13(1):20. doi:10.1186/1471-2199-13-20
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.