Back to Journals » OncoTargets and Therapy » Volume 11
SMAGP a novel biomarker of cervical cancer development and progression
Authors Jia Y, Li H, Liu G, Song F
Received 30 May 2018
Accepted for publication 18 July 2018
Published 15 October 2018 Volume 2018:11 Pages 6925—6935
DOI https://doi.org/10.2147/OTT.S175808
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Yongqin Jia, 1,2 Haiyu Li, 1,3 Geli Liu, 1,2 Fangzhou Song 1,2
1Molecular Medicine and Cancer Research Center, Chongqing Medical University, Chongqing 400016, China; 2Department of Biochemistry and Molecular Biology, Chongqing Medical University, Chongqing 400016, China; 3Department of Infectious Disease, Chongqing Infectious Disease Medical Center, Chongqing 400030, China
Introduction: Cervical cancer, one of the most common malignant gynecological tumors, is a significant burden on the health of females worldwide. The purpose of this study was to investigate genes associated with lymph node metastasis in cervical cancer.
Methods: We report on the lymph node metastasis-associated gene, small cell adhesion glycoprotein (SMAGP), as a key regulator of cervical cancer development and progression. SMAGP expression levels were investigated in 70 cervical squamous cell carcinoma samples and 10 normal cervical squamous epithelium samples.
Results: Immunohistochemistry analysis revealed that SMAGP protein levels were significantly elevated in cervical cancer tissue compared with normal cervical squamous epithelium. Silencing of SMAGP induced cell cycle arrest, inhibited the cell proliferation and colony formation ability of cervical cancer cells in vitro and suppressed their tumorigenic potential in nude mice. In addition, SMAGP knockdown reduced expression of epithelial mesenchymal transition-related proteins, including vimentin, β-cadherin, MMP2, and Twist.
Conclusion: Together, our findings demonstrate that SMAGP plays a critical role in cell proliferation and tumorigenesis and could be a new therapeutic target in cervical cancer.
Keywords: cervical cancer, small cell adhesion glycoprotein, bioinformatics, cell cycle
Erratum for this paper has been published
Introduction
Cervical cancer is the second most common malignancy in women with an estimated 530,000 new cases and 275,000 deaths annually worldwide,1,2 and the majority of female cancer deaths occur in developing countries.3 Various treatments for cervical cancer are available, according to tumor grade and stage, including surgery and radiation therapy. However, treatment for advanced stage cervical cancer remains a public health concern.4,5 In addition, the prognosis of patients with cervical cancer is influenced by numerous factors and not all women with this disease fully recover. Although research into cervical cancer has recently progressed significantly, the condition remains associated with high levels of morbidity and mortality, partly due to a lack of clarity regarding its underlying molecular mechanisms.6 Hence, the identification of novel biomarkers for cervical cancer diagnosis and treatment is required.
As metastasis is the most important process influencing the deterioration of the condition of patients with cancer and leads to the majority of cancer deaths, the inhibition of tumor metastasis is a primary goal for tumor treatment. Small transmembrane and glycosylated protein (SMAGP) consists of 97 amino acids and contains a transmembrane domain.7 SMAGP localizes to the lateral face of the plasma membrane and is expressed in the cytoplasm after cell transformation.8 To understand the functions of SMAGP in cancer progression, we investigated its expression levels in cervical cancer and its effects on the growth and migration of cervical cancer cells. Our findings demonstrate that SMAGP has a critical role in cell proliferation and tumorigenesis and could serve as new therapeutic target in cervical cancer.
Materials and methods
Identification of differently expressed genes (DEGs)
The original data of the GSE26511 data detected by the Affymetrix Human Genome U133 Plus 2 Array, were downloaded from Gene Expression Omnibus database (GEO, http://www.ncbi.nlm.nih.gov/geo/). In total, 20 cervical cancer samples that were lymph node negative and 18 cervical cancer samples that were lymph node positive were used. The raw data were normalized by using R software (https://www.r-project.org/) with Affy (https://www.rdocumentation.org/packages/affy/versions) package (version 3.3.2 John Chambers, NJ, USA), and then differently expressed genes were obtained by using R limma package23 (version 3.28.14. Gordon Smyth. Canberra, Australia). The corresponding P-value was calculated by using the Student’s t-test. Gene expression was considered as significantly different when P-value <0.05 and |log2FC|>1. The Pearson correlation coefficient between mRNA, and mRNA-mRNA co-expression network were constructed by using R software. The Pearson coefficient threshold of the mRNA height correlation in this paper is |correlation|≥0.80.
Cell culture
Cancer cell lines, CaSki, Siha, Hela and C33A, were obtained from the The Cell Bank of Type Culture Collection of Chinese Academy of Sciences. All cell lines were cultured in high glucose DMEM medium (Hyclone, Marlborough, MA, USA) supplement with 10% fetal bovine serum (FBS) (Cell-box, Canberra, Melbourne, VIC, Australia) and 1% penicillin-streptomycin (Beyotime, Shanghai, China) in a 5% CO2 incubator at 37°C. In this study, these cells in exponential growth phase were used.
RNA isolation and quantitative real-time PCR
The total RNA was isolated by using total RNA kit I (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. The quality of RNA was verified, and the concentration of RNA was measured by Nanodrop 2000 (Bio-Rad, Hercules, CA, USA). cDNA was transcript by using a reverse transcription kit (Thermo Fisher Scientific, Waltham, MA, USA) with original 500 ng RNA. Quantitative real-time PCR was performed using SYBR-Green PCR Master Mix (TAKARA Bio Inc.). Primers used in this study were as follows: SMAGP: 5′-GCCTCCTGACTACTCCTTCTC-3′ and 5′-AAGATCACGACCGAGAGCAG-3′; CDK2: 5′-CCAGGAGTTACTTCTATGCCTGA-3′ and 5′-TTCATCCAGGGGAGGTACAAC-3′; CDK4: 5′-ATGGCTACCTCTCGATATGAGC-3′ and 5′-CATTGGGGACTCTCACACTCT-3′; Cyclin E: 5′-CGCTGTTCTGGTCTGGATTG-3′ and 5′-TGGTAGTTGTGTGCGTTTGG-3′; β-actin: 5′-TCCACGAGACCACCTACAAC-3′ and 5′-TCTGGTGGGGCAATGATCTT-3′. The PCR reaction conditions were performed at 95°C for 3 minutes, 40 cycles at 95°C for 5 seconds, 55°C for 30 seconds and 72°C for 30 seconds. The reaction was performed using the SYBR-Green PCR master mix (TAKARA Bio Inc.).
Immunohistochemistry (IHC)
The tissue microarray (AURAGENE, Shanghai, China) used in this study containing 25 cases in early stage, 45 cases in advanced stage and 10 cases of normal tissue. Immunohistochemical experiments were performed according to the manufacturer’s instructions (Beijing Zhongshan Jinqiao Biological Technology Company Limited, Beijing, China). In detail, dewax by dimethylbenzene twice after placing the chip under 50°C for 2 hours, and use 100%, 95%, 85%, 75% ethanol to hydrate at 5 min intervals. Then the chip was incubated with SMAGP antigen at 95°C for 20 minutes in the sodium citrate buffer before incubation with hydrogen peroxide for 10 minutes, and then it was washed with PBST (PBS with 0.1% Tween-20) three times. Then the chip was incubated with rabbit polyclonal anti-SMAGP (Bioss, Beijing, China; 1:400) at 4°C overnight before incubated with a secondary antibody for 2 hours at room temperature. Followed by hematoxylin staining, 3, 3′-diaminobenzidine (DAB) color reagent was added. Finally the chip was dehydrated and then installed in neutral resins.
Immunofluorescence (IF)
Before cells were seeded in plate and cultured overnight, sterile cover slips were placed in the 24-well plate. Cells were immobilized using 4% polyoxymethylene for 20 minutes and washed with PBS. After cells were permeabilized by 0.5% Triton X-100, cells were blocked with 10% goat serum for 30 minutes. After that, cells were incubated with rabbit polyclonal anti-SMAGP (Bioss, 1:500) overnight at 4°C, then the secondary anti-rabbit was added with FITC fluorescence and the cells were incubated for a further 2 hours at room temperature under light avoidance conditions. Finally, nuclear staining was performed with delta-interacting protein A. The expression and location of SMAGP was visualized using a fluorescence microscope.
RNA interference and lentiviral transfection
All siRNA and lentiviral particles used in this study were chemically synthesized by GenePhama (Shanghai, China). The siRNA sequences are as follows: si299: 5′-GCACAGCACUCAUUGCAGUTT-3′ and 5′-ACUGCAAUGAGUGCUGUGCTT-3′; si440: 5′-UCCAGAUGAGAGUGACUUTT-3′ and 5′-AAGUCACUCUCCAUCUGGATT-3′; si581: 3′-GGGAGAAGCAUUGAUUGAUTT-3′ and 5′-AUCAAUCAAUGCUUCUCCCTT-3′. The negative control sequence is: 5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′. Transfection efficiency was validated at 48 hours after transfection. Cells were transfected with siRNA oligonucleotide by RNAi MAX (Thermo Fisher Scientific, Waltham, MA, USA). Lentiviral particles were transducted to CaSki cells to establish a stable cell line, the sequence of sh-SMAGP are: 3′-GGGAGAAGCAUUGAUUGAUTT-3′ and 5′-AUCAAUCAAUGCUUCUCCCTT-3′; shNC: 5′-TTCTCCGAACGTGTCACGT-3′. Twenty-four hours after transduction, 3 μg/mL purinamycin was added to the medium to obtain stable cell line.
Cell counting kit-8 (CCK8)
The CaSki cells (100 μL/well) were inoculated to 96-well plate 48 hours after siRNA transfection and separated into two groups; siNC and si581, shNC and shSMAGP. CCK8 reagent (10 μL/well) was then added and the cells were incubated in a 5% CO2 incubator at 37°C for 1 hour. The absorbance was measured at a wavelength of 450 nm every 24 hours for 5 days.
EdU assay
CaSki cells were seeded into a 24-well plate with sterile cover slips and cultured overnight. Seventy-two hours after siRNA (siNC and si581) transfection, cells were immobilized in 4% polyoxymethylene and stained with 100 uL 1X Apollo® dyeing reaction liquid (Ribobio Co Ltd., Guangzhou, China). Finally, photos were obtained using a fluorescence microscope and preserved.
Colony formation assay
Cells (1,000 cells/well) transfect with siRNA were inoculated in a 6-well plate and cultured in a 5% CO2 incubator at 37°C for 2 weeks with high glucose DMEM medium. Clones that appeared to be visible to the naked eyes were washed by PBS three times, immobilized in 4% polyoxymethylene for 20 minutes, and stained with 0.5% crystal violet for 20 minutes, and washed with PBS again. Every colony with more than 50 cells was counted under microscope.
Flow cytometry analysis
Seventy-two hours after siRNA transfection, cells were diluted to a density of 1–5×105/mL with 70% ethanol and kept at 4°C overnight. Then the cells were analyzed by flow cytometry.
Migration and invasion assays
Cell migration activity was assessed by a wound healing and transwell assay. About 6×105 cells transfect with siRNA were seeded into a 6-well plate and cultured overnight. The cells were scratched with a 200 μL pipette tip. Floating cells were washed with PBS, and then the cells were observed and imaged under a microscope at different time-points. The distance of the uncovered wound gap was quantified using ImageJ software (version: 1.8.0), the relative wound closure was assessed at 36 hours relative to 0 hour. For the cell migration assay, cells transfected with siRNA (2–3×104) were resuspended in 200 μL serum-free medium and seeded into the upper chamber, 500 μL complete medium was added to the lower chamber as chemotactic factor. For the transwell assay, 3×104 cells transfected with siRNA were seeded in transwell chamber, after 24 hours the cells that had migrated into lower surface were fixed and stained with 0.5% crystal violet. Invasion was based on the migration experiment, cells were seeded into a Matrigel-coated transwell chamber. Each experiment was performed at least three times.
Western blotting
All cultured cells were lysed in proper volume with RIPA lysis buffer containing 1% phenylmethanesulfonyl fluoride and kept on the ice for 30 minutes, followed by centrifugation at 12,000 rpm at 4°C for 15 minutes. Supernatant was collected, and the concentration was determined by the bicinchoninic acid protein assay. The loading buffer (Beyotime) was added to supernatant and protein was denatured at 100°C for 10 minutes. After separated in SDS-PAGE gels (Beyotime), the protein was transferred to a PVDF membrane. The membrane was blocked in TBST containing 5% skim milk at room temperature for 2 hours, then incubated with primary antibody solution overnight at 4°C before incubation with an appropriate secondary antibody for 2 hours at room temperature. The protein expression was detected using the ECL kit (Advansta, San Jose, CA, USA).
Tumor metastasis assay in vivo
Approval for the use of the nude mice was obtained from the ethical committee of the Chongqing Medical University (Yuzhong, Chongqing, China), and all animal experiments were carried out in accordance with the National Institutes of Health Guidelines for Animal Research (Guide for the Care and Use of Laboratory Animals). Special care was taken to minimize the suffering of animals. Hence, all experiments were performed following relevant institutional and national guidelines and regulations. All female nude mice were purchased from an animal facility of the Chongqing Medical University. Cells with stable SMAGP siRNA transfection (shNC, shSMAGP) were injected intraperitoneally to nude mice. At 28 days after injection, the tumor mass and related tissues were removed, photo’s were taken and specimens preserved for further analysis.
Statistical analysis
All statistical data were analyzed by SPSS statistical software (IBM Corporation, Armonk, NY, USA), data were presented as a mean ± SD, Student’s t-test and one-way ANOVA were used for data analysis. The ImageJ was used to analysis results of immunohistochemistry, P<0.05 was considered statistically significant.
Results
Identification of SMAGP as a prognostic gene
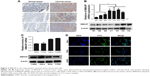
A bioinformatics method was used to screen for SMAGP, and the results showed that SMAGP was highly expressed in cervical cancer. In total, 38 samples including 20 normal tissues and 18 cervical cancers were used for the informatics study. After differently expressed genes were selected, 300 genes including 92 up-regulated genes and 208 down regulated genes were obtained. A heat map of hierarchical cluster analysis showed expression of genes differed between cervical cancer and normal tissues (Figure 1A). A co-expression network diagram of mRNA was established (Figure 1B). Finally, the expression levels of SMAGP in various tumors were compared (Figure 1C), and the results showed that SMAGP was significantly up-regulated in cervical cancer. These results prompted the selection of SMAGP in cervical cancer for further investigation.
SMAGP is up-regulated in cervical cancer tissues and cell lines
The dataset GSE26511 was downloaded from GEO to screen for the up-regulated genes in cervical cancer, and mRNA-mRNA co-expression network map was constructed. DAVID, KEGG PATHWAY and other bioinformatics tools were used to analyze genes that were significantly expressed. Finally, we selected SMAGP and compared the expression levels of SMAGP in different tumors. To different SMAGP expression in cervical squamous cell carcinoma tissues and normal cervical squamous epithelium tissues, we examined the expression of SMAGP in 70 cases of cancer tissue and 10 normal controls by immunohistochemistry (Table 1). The results showed that SMAGP expression was increased in 43 of the 70 (61.4%) cervical carcinoma tissues and was not expressed in normal tissues, which indicates that SMAGP expression level is higher in cervical carcinoma tissues compared to normal tissues (Figure 2A and B). We also found that SMAGP is mainly expressed in the membranes in non-cancerous tissues, whereas, SMAGP is mainly expressed in the cytoplasm in cervical carcinoma tissues. Given that SMAGP encodes small cell adhesion glycoprotein, we hypothesized that SMAGP may play an important role in tumorigenesis of cervical cancer. Then, we examined the expression levels of SMAGP in four cervical cancer cell lines including SiHa, HeLa, CaSki and C33A (Figure 2C and D). The results showed that SMAGP was up-regulated in cervical cancer, and SMAGP was highly expressed in Hela and CaSki, locates in both the cell membrane and cytoplasm.
SMAGP knockdown inhibited cells proliferation by blocking cell cycle at G0/S phase
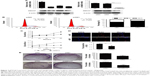
To evaluate the functions of SMAGP in cervical cancer, a SMAGP knockdown cell model was constructed by transfecting SMAGP siRNA into CaSki cells. CaSki cells were transfected with siRNA (siNC, si299, si440, si581) and lentivirus (shNC, shSMAGP). The expression levels of SMAGP were examined by RT-PCR and Western blotting at mRNA and protein levels after 48 hours and 72 hours transfection respectively (Figure 3A). There results showed that siRNA and lentivirus could silence the expression of SMAGP. As shown in figure, sequence si581 largely reduced SMAGP expression, thus sequence si581 was selected for further experiments.
To investigate the effect of SMAGP on the proliferation and viability of the cervical cancer cell line CaSki, flow cytometry, Cell Counting Kit-8 (CCK8), colony formation assays and EdU assays were applied. After 72 hours siRNA transfection, flow cytometry results showed that SMAGP knockdown blocked cell cycle at G0/S phase (Figure 3B). SMAGP knockdown inhibited cells proliferation and viability compared with the control cells as shown by the CCK8, colony formation and EdU assays (Figure 3C–E).
Knockdown SMAGP inhibited cell migration and invasion
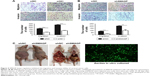
The migration and invasion ability of CaSki cells were quantified by the transwell assay. Transwell assay results suggested that silenced SMAGP expression might inhibit the migration and invasion of cells (Figure 4A). The parallel migration and invasion abilities of CaSki cells with stable SMAGP expression were investigated. Consistent results were achieved with the two assays (Figure 4B). These results indicated that the migration and invasion ability of cervical cancer cells was significantly decreased in CaSki cells transfected with siRNA.
Knockdown SMAGP inhibited CaSki metastasis in nude mice
In order to evaluate the effects of SMAGP in vivo, CaSki cells with stable shSMAGP expression were injected into nude mice. At 28 days after injection, the mice were euthanized, and the tumor mass and related tissues were removed and used for further analysis. Obvious swelling in the abdominal cavity of nude mice was found in the control group, and the ascites was collected and cultured in a culture dish. Green fluorescence was observed, and the cell morphology was consistent with the injected CaSki cells. In the control group, a large number of tumors were found around the intestine, but tumors only developed at the injection site in the shSMAGP group (Figure 4C). The results revealed that knockdown SMAGP inhibited CaSki cell migration to the intestines of the nude mice.
Knockdown SMAGP inhibited the expression of cyclin and epithelial mesenchymal transmission related protein in CaSki cells
The cell cycle plays an important role in the process of cell proliferation, and cell proliferation inhibition is an important method to treatment early stage tumors. On the other hand, metastasis and invasion are the main factors of cancer deterioration, and epithelial mesenchymal transmission (EMT) happened in the early stage of epithelial cell tumor metastasis, which is one of the important mechanisms for malignant tumor cells to obtain migration and invasion abilities.9 To investigate the possible mechanisms that SMAGP plays in cell cycle and metastasis, knockdown of SMAGP was achieved and the expression of CDK2, CDK4, cyclin E and EMT related proteins were examined using RT-PCR and Western blotting. The results showed that CDK2, CDK4 and Cyclin E were down regulated after SMAGP knockdown in the cell cycle (Figure 5A). Vimentin, β-cadherin, MMP2, Twist were down regulated in EMT (Figure 5B).
Discussion
Cervical cancer is one of the most serious malignant tumors, and is associated with high rates of morbidity and mortality, particularly in developing countries, including China.10,11 Worldwide, a large number of new cervical cancer cases are diagnosed every year, and more than half of patients diagnosed died from the cancer.3 Tumor progression involves tumor cell proliferation and metastasis and novel targeted therapeutic strategies are urgently required for patients with cervical cancer.
In this study, expression levels of SMAGP in cervical cancer cells were determined by real-time PCR and Western blotting and demonstrated that SMAGP is highly expressed in the HeLa and CaSki cell lines. CaSki cells are a human cervical cancer bowel metastatic cell line. Our results demonstrated that SMAGP expression levels are associated with lymph node-positive cervical cancer.
In this study, we aimed to investigate the functions of SMAGP in the cell cycle and metastases in cervical cancer. Our results reveal that the expression levels of genes encoding proteins involved in the cell cycle, including CDK2, CDK4, and Cyclin E reduced, while factors participated in EMT (β-cadherin, vimentin, and MMP2) reduced. Suppression of CDK2 and CDK4 expression can induce G1 phase arrest12 and EMT is a complex biological process that has a critical role in embryonic development, as well as tumor metastasis and progression;13–15 however, these mechanisms remain unclear and require further exploration.
SMAGP has high expression levels in various cancers, such as colon, liver, lung and breast7 and can bind protein 4.1 via an intracellular binding domain. Protein 4.1 is a cytoskeletal protein encoded by of a group of paralogous genes, including EPB41 (4.1R), EPB41L2 (4.1G), EPB41L3 (4.1B) and EPB41L1 (4.1N).16–19 Further exploration of the mechanism of action underlying the interaction between SMAGP and protein 4.1 may identify protein 4.1 as another potential therapeutic target in cervical cancer. The SMAGP protein intracellular domain can also bind to membrane-associated guanylate kinase (MAGUK), protein P55 and CASK. MAGUKs are a family of scaffold proteins that are enriched at cellular junctions and are important for cell adhesion and intercellular signal transduction.20–22 Our data indicate that SMAGP is associated with tumor metastases and may be a key factor that inhibits tumor deterioration; however, SMAGP extracellular binding proteins remain unidentified and require further exploration. Nevertheless, SMAGP is a potential therapeutic target molecule. Further studies are required to verify the regulatory mechanisms that mediate the interactions between SMAGP and molecules involved in cell proliferation, cell migration, and the invasion of cervical cancer cells.
Author contributions
Experiments performance and data collection: JY, LH. Data analysis: LG. Reagents/materials Contribution: GL. Paper writing: JY, LH. Paper revision: SF. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Li F, Wang Z, Lu G. TRIM28 promotes cervical cancer growth through the mTOR signaling pathway. Oncol Rep. 2018;39(4):1860–1866. | ||
Tian R, Li X, Gao Y, Li Y, Yang P, Wang K. Identification and validation of the role of matrix metalloproteinase-1 in cervical cancer. Int J Oncol. 2018;52(4):1198–1208. | ||
Liao S, Xiao S, Chen H, et al. The receptor for activated protein kinase C promotes cell growth, invasion and migration in cervical cancer. Int J Oncol. 2017;51(5):1497–1507. | ||
Dai B, Yang T, Ma Y, et al. HMQ-T-F2 exert antitumour effects by upregulation of Axin in human cervical HeLa cells. J Cell Mol Med. 2018;22(5):2955–2959. | ||
Yang W, Tan W, Zheng J, Zhang B, Li H, Li X. MEHP promotes the proliferation of cervical cancer via GPER mediated activation of Akt. Eur J Pharmacol. 2018;824:11–16. | ||
Liu P, Ma S, Liu H, Han H, Wang S. HCFU inhibits cervical cancer cells growth and metastasis by inactivating Wnt/β-catenin pathway. J Cell Biochem. In press 2017. | ||
Tarbé NG, Rio MC, Weidle UH. SMAGP, a new small trans-membrane glycoprotein altered in cancer. Oncogene. 2004;23(19):3395–3403. | ||
Tarbé NG, Rio MC, Hummel S, Weidle UH, Zöller M. Overexpression of the small transmembrane and glycosylated protein SMAGP supports metastasis formation of a rat pancreatic adenocarcinoma line. Int J Cancer. 2005;117(6):913–922. | ||
Yue QY, Zhang Y. Effects of Linc00460 on cell migration and invasion through regulating epithelial-mesenchymal transition (EMT) in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22(4):1003. | ||
Liu M, Jia J, Wang X, Liu Y, Wang C, Fan R. Long non-coding RNA HOTAIR promotes cervical cancer progression through regulating BCL2 via targeting miR-143-3p. Cancer Biol Ther. 2018;19(5):1–9. | ||
Shi X, Ran L, Liu Y, et al. Knockdown of hnRNP A2/B1 inhibits cell proliferation, invasion and cell cycle triggering apoptosis in cervical cancer via PI3K/AKT signaling pathway. Oncol Rep. 2018;39(3):939–950. | ||
Chen MJ, Cheng AC, Lee MF, Hsu YC. Simvastatin induces G1 arrest by up-regulating GSK3β and down-regulating CDK4/cyclin D1 and CDK2/cyclin E1 in human primary colorectal cancer cells. J Cell Physiol. 2018;233(6):4618–4625. | ||
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. J Clin Invest. 2009;119(6):1438. | ||
Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14(6):818–829. | ||
Yuan H, Kajiyama H, Ito S, et al. ALX1 induces snail expression to promote epithelial-to-mesenchymal transition and invasion of ovarian cancer cells. Cancer Res. 2013;73(5):1581–1590. | ||
Parra M, Gascard P, Walensky LD, Snyder SH, Mohandas N, Conboy JG. Cloning and characterization of 4.1G (EPB41L2), a new member of the skeletal protein 4.1 (EPB41) gene family. Genomics. 1998;49(2):298–306. | ||
Rangel L, Lospitao E, Ruiz-Sáenz A, Alonso MA, Correas I. Alternative polyadenylation in a family of paralogous EPB41 genes generates protein 4.1 diversity. RNA Biol. 2017;14(2):236–244. | ||
Tang TK, Leto TL, Correas I, Alonso MA, Marchesi VT, Benz EJ. Selective expression of an erythroid-specific isoform of protein 4.1. Proc Natl Acad Sci U S A. 1988;85(11):3713–3717. | ||
Walensky LD, Blackshaw S, Liao D, et al. A Novel Neuron-Enriched Homolog of the Erythrocyte Membrane Cytoskeletal Protein 4.1. J Neurosci. 1999;19(15):6457–6467. | ||
Oliva C, Escobedo P, Astorga C, Molina C, Sierralta J. Role of the maguk protein family in synapse formation and function. Dev Neurobiol. 2012;72(1):57–72. | ||
Ye F, Zeng M, Zhang M. Mechanisms of MAGUK-mediated cellular junctional complex organization. Curr Opin Struct Biol. 2018;48:6–15. | ||
Zhu J, Shang Y, Zhang M. Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nat Rev Neurosci. 2016;17(4):209–223. | ||
Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3(1):1–25. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.