Back to Journals » International Journal of General Medicine » Volume 15
SIRT7 is a Prognostic Biomarker in Kidney Renal Clear Cell Carcinoma That is Correlated with Immune Cell Infiltration
Authors Zhang W , Qian Y, Jin X, Wang Y, Mu L, Jiang Z
Received 21 December 2021
Accepted for publication 10 March 2022
Published 19 March 2022 Volume 2022:15 Pages 3167—3182
DOI https://doi.org/10.2147/IJGM.S353610
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Wei Zhang,1 Yue Qian,1 Xue Jin,2 Yixian Wang,1 Lili Mu,1 Zhixia Jiang2
1Department of Pathogen Biology, Guizhou Nursing Vocational College, Guiyang, People’s Republic of China; 2Guizhou Nursing Vocational College, Guiyang, People’s Republic of China
Correspondence: Wei Zhang, Department of Pathogen Biology, Guizhou Nursing Vocational College, Guiyang, 550000, Guizhou, People’s Republic of China, Email [email protected]
Background: SIRT7 has been shown to be expressed in many cancer types, including clear cell renal cell carcinoma (KIRC), but its functional role in this oncogenic context remains to be firmly defined. This study was designed to explore correlations between SIRT7 and KIRC characteristics using the TCGA database.
Methods: Relationships between SIRT7 expression and KIRC patient clinicopathological characteristics were assessed through Kruskal–Wallis tests, Wilcoxon signed-rank tests, and logistic regression analyses. Area under the ROC curve (AUC) values were used to assess the prognostic value of SIRT7 as a means of classifying clear cell renal cell carcinoma patients. The functional role of SIRT7 in this cancer type was assessed through GO/KEGG enrichment analyses and immune cell infiltration analyses.
Results: In KIRC patients, higher levels of SIRT7 expression were associated with Race, M stage, T stage (all P < 0.05). SIRT7 offered significant diagnostic value in ROC curve analyses (AUC = 0.912), and elevated SIRT7 levels were linked to worse patient overall survival (OS; P < 0.001). The expression of SIRT7 was independently related with KIRC patient OS (HR: 1.827; 95% CI: 1.346– 2.481; P< 0.001). In GO/KEGG analyses, SIRT7 was found to be associated with ubiquitin-mediated proteolysis and nucleotide excision repair. Higher SIRT7 expression was related to the enhanced infiltration of certain immune cells.
Conclusion: Increased SIRT7 expression was associated with a worse KIRC patient prognosis, and immune infiltrates, suggesting it may offer value as a prognostic biomarker for this cancer type.
Keywords: SIRT7, biomarker, KIRC, prognosis, tumor-immune infiltration
Introduction
Renal cell carcinoma (RCC) is the deadliest form of urological cancer.1 Between 60% and 85% of all RCC cases are of the kidney RCC (KIRC) subtype, with approximately 1/3 of KIRC patients exhibiting metastases at the time of diagnosis that are associated with poor survival outcomes.2 KIRC tumors are highly heterogeneous in nature, often harboring myriad complicated genetic alterations and signaling abnormalities. A variety of genetic, environmental, and lifestyle-related factors are linked to the etiology of this tumor type. The primary current treatments for KIRC patients include surgery, chemotherapy, radiotherapy, and targeted treatment. However, the results of clinical trials testing targeted molecular therapeutics have been unsatisfactory to date.3,4 Recent efforts based on studies of the Biology of KIRC have identified novel therapeutic targets with the potential to improve patient outcomes. Multiple signaling pathways have been shown to serve important functional roles in the regulation of KIRC onset, progression, recurrence, and therapeutic resistance, yet the underlying molecular mechanisms remain to be fully elucidated.5,6 Identifying reliable, sensitive biomarkers of KIRC has the potential to aid in the diagnosis and prognostic evaluation of patients, enabling clinicians to better guide patient care and to formulate appropriate individualized treatment regimens.
Sirtuins (SIRTs) are a family of 7 (SIRT1-7) highly-conserved class III nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylases.7 These SIRTs localize to different subcellular regions and exhibit distinct catalytic activities and substrates, regulating cellular survival, transcription, DNA repair, differentiation, proliferation, longevity, senescence, and other biological processes.8–10 SIRT7 was the most recently designated SIRT family protein. Encoded on chromosome 17q25.3, SIRT7 has been shown to serve as a key regulator of rDNA transcription, processing, and maturation.11 A growing body of evidence has also shown that SIRT7 plays important roles in tumor cells wherein it can shape central oncogenic processes such as metastasis, proliferation, transformation, radioresistance, and chromatin modification-related stress responses.12 In thyroid cancer, SIRT7 can activate p70S5K1 and AKT via the DBC1/SIRT1 axis.13 Furthermore, SIRT7 can upregulate p-ERK and p-MEK expression at the epigenetic level, thereby promoting the progression of colorectal cancer.14 In hepatocellular carcinoma (HCC) cells, the expression of SIRT7 has been reported to fall under the control of miR-145,15 while in prostate cancer cells it can drive enhanced migratory and proliferative activity.16 Intriguingly, SIRT7 plays an important role in the development and progression of human bladder cancer.17 However, there is currently insufficient data available regarding the biological importance of SIRT7 in KIRC.
The present study was formulated to establish the relative expression of SIRT7 in KIRC patient tumor tissues and healthy control tissues based upon RNA-sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA) and GTEx databases. The relationship between the expression of SIRT7 and KIRC patient clinicopathological features was then assessed, and the prognostic role of this SIRT family protein in this cancer type was examined. To uncover the potential functions of SIRT7, a gene set enrichment analysis (GSEA) approach was employed. Lastly, we herein conducted a thorough investigation of the possible mechanisms whereby SIRT7 controls KIRC onset and progression by examining associations between the expression of this gene and immune cell infiltration.
Methods
Datasets
Clinical data and RNA-seq results pertaining to 539 KIRC patients and 72 paired paracancerous control tissue samples were downloaded from the TCGA database in the level 3 FPKM format. These data were then converted into the TPM format for additional use. In addition, RNA-seq data were downloaded from the TCGA and the GTEx databases from UCSC Xena in the TPM format.18 All analyses were consistent with the Helsinki Declaration (as revised in 2013). SIRT7 mRNA expression was also assessed in the GSE563757 and GSE40435 GEO datasets.
TIMER Database Analyses
The TIMER database enables users to study the differential expression between tumor and adjacent normal tissues for any gene of interest across all TCGA tumors.19,20 For this study, the TIMER database was instead used to assess SIRT7 expression in different cancers.
The Human Protein Atlas
The Human Protein Atlas compiles transcriptomic and proteomic data pertaining to different cellular and tissue samples, including 44 healthy tissues and 20 different types of cancer. Immunohistochemistry results relevant to KIRC were downloaded from this database.
SIRT7-Related Gene Enrichment Analysis
SIRT7-interacting proteins were identified using the STRING database to identify predicted interaction partners, with the species being set to Homo sapiens. Parameters for the prediction of interacting proteins were as follows: meaning of network edges (“evidence”), Low confidence (0.150), active interaction sources (“experiments”), and the maximum number of interactors to display (“no more than 50”). The GEPIA2.0 “Similar Gene Detection” module was also utilized to predict the top 100 SIRT-associated genes in healthy tissue and TCGA tumor tissue datasets. These results were then integrated for KEGG pathway analyses, and enriched pathways were interpreted with the “tidyr” and “ggplot2” R packages. Gene Ontology (GO) analyses were conducted with the R “clusterProfiler” package. The enriched biological processes, molecular functions, and cellular components (BPs, MFs, CCs) were represented with cnet-plots. R v 3.6.3 was used for all analyses, with a two-tailed P < 0.05 as the significance threshold.
Immune Infiltration Analysis
To approximate immune cell infiltration in KIRC tumors based on gene expression data, a single sample GSEA (ssGSEA) analysis was conducted using the GSVA v 3.6 R package. Tumor infiltration by 24 different immune cell types was estimated based on specific gene expression profiles.21 Correlations between SIRT7 expression and infiltration by these different immune cell subsets were assessed through Spearman and Wilcoxon rank-sum tests, while Pearson correlation analyses were utilized to assess the relationship between SIRT7 expression and the expression of different immune checkpoint genes. The relationship between SIRT7 expression and immune cell migratory activity was evaluated by analyzing chemokines/chemokine receptors with the “chemokine” TISIDB database module.
TISIDB
The TISIDB website enables analyses of the relationships between gene expression and tumor immune cell infiltration. This website was used to assess the expression of SIRT7 in the context of different immune tumor subtypes including the C1 (wound healing), C2 (IFN-γ-dominant), C3 (inflammatory), C4 (lymphocyte-depleted), C5 (immunologically quiet), and C6 (TGF-β-dominant) subtypes based on the expression of the SIRT7 gene in different molecular subtypes included in the TCGA database.
Statistical Analysis
R v. 3.6.3 was used for statistical testing. Relationships between SIRT7 expression and patient clinicopathological characteristics were assessed via chi-squared tests, logistic regression analysis, Fisher’s exact test, and Wilcoxon rank-sum tests (WRST). Associations between SIRT7 expression and the overall survival (OS) of KIRC patients in the TCGA database were assessed via Kaplan-Meier analyses. Univariate and multivariate analyses were conducted to evaluate correlations between clinical and genetic findings, and Cox proportional hazard models were used to evaluate patient OS. P < 0.05 was the significance threshold.
Results
Baseline Patient Characteristics
In total, data from 539 patients with KIRC in the TCGA database were downloaded in March 2021 (Table 1). These KIRC patients were then stratified into two groups based upon whether patients were above or below the mean SIRT7 expression levels (n=270 and n=269, respectively). Next, associations between SIRT7 levels and KIRC patient clinicopathological properties were assessed, revealing the expression of this gene to be related to T stage, M stage, and Race (P < 0.05).
 |
Table 1 Correlation Between SIRT7 Expression and Clinicopathological Characteristics in KIRC |
SIRT7 Were Highly Expressed in KIRC
To explore the potential functional roles of SIRT7 in KIRC, we began by using the TIMER database to survey the expression of this gene in patient samples. This analysis indicated that SIRT7 was expressed at significantly higher levels in KIRC tumors as compared to normal renal tissue samples (Figure 1A), and that it was similarly expressed at high levels in other cancer types including bladder urothelial cancer, breast invasive carcinoma, cholangiocarcinoma, esophageal carcinoma, kidney renal papillary cell carcinoma, lung adenocarcinoma, prostate adenocarcinoma, HCC, and stomach adenocarcinoma tissues relative to corresponding adjacent control tissues. SIRT7 levels were, in contrast, lower in colon adenocarcinoma and kidney chromophobe tissues relative to adjacent control tissues. No data pertaining to SIRT7 expression was available for adrenocortical carcinoma, lymphoid neoplasm diffuse large B-cell lymphoma, or brain lower grade glioma tumors in this database.
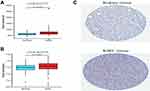
To explore the link between SIRT7 expression and KIRC patient clinical findings, we leveraged the TCGA database and found that, consistent with the above results, SIRT7 was significantly upregulated in 539 KIRC tumor tissue samples as compared to 72 normal control tissue samples (P<0.001; Figure 1B). Meanwhile, we also evaluated the SIRT7 expression in 72 KIRC tissues and their matched adjacent tissues, and the obtained results indicated that the SIRT7 expression was elevated in KIRC tissues (P <0.001;Figure 1C). The GTEx and TCGA databases were then combined to explore SIRT7 mRNA expression patterns in KIRC, revealing it to again be significantly upregulated in 531 tumor tissue samples relative to 100 normal tissue samples (P <0.001; Figure 1D). This was similarly confirmed in an analysis of the GEO database (P<0.001, 0.005; Figure 2A and B). Correspondingly, the expression of SIRT7 protein is significantly higher in KIRC tissue as compared to normal tissue in comparison to that in normal tissue in the Human Protein Atlas (Figure 2C). Together, these data highlighted a potential role for SIRT7 as a regulator of KIRC onset and progression.
SIRT7 Upregulation is Associated with Poor KIRC Clinical Features
In total, clinical information and SIRT7 expression levels were available for 539 KIRC patients in the TCGA database. We therefore explored the relationship between SIRT7 levels and these clinicopathological findings through univariate analyses which revealed SIRT7 expression to be associated with Race, T stage, M stage, and pathologic stage (Table 2). Specifically, elevated SIRT7 levels were associated with T stage, M stage, Race, and pathologic stage (all P< 0.05; Figure 3A–D). These results suggested that KIRC patients with higher SIRT7 levels are more likely to exhibit more advanced disease as compared to patients with lower levels of SIRT7 expression.
 |
Table 2 The Expression of SIRT7 Has Been Linked with the Clinicopathological Features (Logistic Regression) |
Elevated SIRT7 Expression is Independently Associated with Decreased KIRC Patient Overall Survival
The relationship between KIRC patient OS and SIRT7 expression levels was next assessed via the Kaplan-Meier approach. This revealed higher levels of SIRT7 to be associated with a worse prognosis (P<0.001; Figure 4A). Subgroup analyses further confirmed a link between SIRT7 upregulation and a poorer prognosis for KIRC patients with T2 stage (P=0.015), T3 stage (P=0.04), N0 stage (P<0.001), M0 stage (P=0.037), Male (P<0.001), Histologic grade G3 (P=0.001), Histologic grade G4 (P=0.048), Pathologic stage II/III (P = 0.015), and Pathologic stage IV disease (P = 0.014), as well as for patients > 60 years old (P = 0.004), and ≤ 60 years old (P = 0.003) (Figure 4B–L). SIRT7 is thus an independent predictor of impaired survival among KIRC patients.
Variables significantly related with patient outcomes in univariate Cox regression analyses included T stage, M stage, N stage, pathologic stage, age, Gender Male, histologic grade, and SIRT7 expression levels (all P<0.001). No significant association was found between SIRT7 expression and a poorer prognosis for KIRC patients with T1 stage, T4 stage, N1 stage, Gender Female, Histologic grade G1, Histologic grade G2, and Pathologic stage I (Supplementary Figure S1A–G). In a subsequent multivariate analysis, M stage (P<0.001), SIRT7 expression (P=0.002), pathologic stage (P=0.017), age (P=0.009), and histologic grade (P=0.015) were all identified as independent predictors of KIRC patient OS (Table 3).
 |
Table 3 The Association Between KIRC Patient OS and Clinicopathological Findings Identified Through Univariate and Multivariate Cox Regression Analyses of TCGA Patients |
SIRT7 Offers Value as a Diagnostic Biomarker for KIRC
Next, we employed ROC curves to explore the value of SIRT7 as a diagnostic biomarker for KIRC. The AUC value for SIRT7 was 0.912, consistent with its utility as a gene that can reliably differentiate between KIRC tumors and healthy normal tissue samples (Figure 5A). SIRT7 was similarly reliable as a biomarker capable of differentiating between tumors of different stages, with respective AUC values of 0.912, 0.854, 0.924, and 0.945 for stage I, II, III, and IV KIRC tumors (Figure 5B–E). As subgroup analyses additionally confirmed the diagnostic relevance of SIRT7 for different KIRC patient clinicopathological characteristics above, a nomogram integrating SIRT7 expression and other clinical variables was generated to predict the 1-, 3-, and 5-year OS of KIRC patients (Figure 6).
Analysis of SIRT7-Associated Genes and Proteins in KIRC
In an effort to elucidate the mechanisms whereby SIRT7 functions in KIRC tumors, we next identified predicted SIRT7-interacting proteins and genes. The STRING tool identified 50 putative SIRT7-interacting proteins for which experimental evidence of interactions was available (Figure 7A). After integrating KIRC expression data using the GEPIA2.0 tool, the top 100 genes associated with SIRT7 expression were identified (Figure 7B). This analysis revealed SIRT7 expression to be positively correlated with that of the ENTHD2 (Adaptor Related Protein Complex 4 Accessory Protein 2) (R=0.80), HDAC10 (Histone Deacetylase 10) (R=0.76), SRRT (Serrate, RNA Effector Molecule) (R=0.75), NUP85 (Nucleoporin 85) (R=0.75), ARHGEF1 (Rho Guanine Nucleotide Exchange Factor 1) (R=0.75), and ANKRD13D (Ankyrin Repeat Domain 13D) (R=0.75) genes (all P<0.001). KEGG and GO enrichment analyses were then conducted for these genes by merged the two datasets, with enriched pathways including the ubiquitin-mediated proteolysis and nucleotide excision repair pathways (Figure 7C), suggesting a link between these mechanisms and SIRT7-mediated regulation of tumorigenesis. GO enrichment analyses suggested these genes to be related to nucleotide excision repair, covalent chromatin modification, heterochromatin, DNA damage recognition, and ubiquitin-dependent protein binding (Figure 7C).
The Association Between SIRT7 Expression and KIRC Tumor Immune Cell Infiltration
The infiltration of the tumor microenvironment (TME) by different immune cell populations can shape patient survival outcomes and predict patient prognosis. We thus employed the ssGSEA analysis to assess the link between SIRT7 expression and such immune infiltration in KIRC tumors, revealing SIRT7 levels to be negatively correlated with infiltration by gd T cells, Mast cells, iDCs, Macrophages, Neutrophils, and Th2 cells, as well as positively correlated with infiltration by NK CD56bright cells, T helper cells, Cytotoxic cells, CD8+ T cells, Treg cells, and T cells (all P < 0.05; Figure 8A–G, Supplementary Figure S2).
Based on immune molecular typing profiles in the TISIDB database, we assessed SIRT7 mRNA levels in different tumor immune subtypes and detected significant differences in SIRT7 expression among the C1 (wound healing), C2 (IFN-γdominant), C3 (inflammatory), C4 (lymphocyte-depleted), C5 (immunologically quiet), and C6 (TGF-β dominant) KIRC subtypes (Figure 8H). The significant change of immune cell infiltration level under various copy numbers of SIRT7 in KIRC was observed by using TIMER database (Figure 8I).
Immune checkpoint inhibitors have emerged as increasingly promising tools for the immunotherapeutic treatment of many cancer types.22,23 We thus assessed correlations between SIRT7 expression and the levels of 40 different immune checkpoint genes in KIRC, revealing it to be correlated with 36 of these genes including CTLA4, CD274, CD276, and TNFRSF25 (Figure 9A). CTLA4 is a key biomarker of immune checkpoint inhibition that is amenable to therapeutic targeting.24 Given these observed correlations, these results suggested that SIRT7 may play a central role in modulating the immune responsiveness of KIRC tumors. To more fully explore the link between the expression of SIRT7 and the migratory activity of immune cells, we additionally examined correlations between SIRT7 levels and the expression of different chemokines and chemokine receptors (Figure 9B–G). This revealed SIRT7 to be positively correlated with the expression of CCL5 (r = 0.423, P <2.2e−16), CCL17 (r = 0.302, P =1.35e−12), CXCL2 (r = 0.271, P =2.2e−10), CXCL13 (r = 0.323, P = 2.74e−14), XCL1 (r = 0.373, P<2.2e−16), and XCL2 (r = 0.425, P<2.2e−16) in KIRC. Higher SIRT7 expression may thus be related to the enhanced infiltration of certain immune cells into the TME.
Discussion
SIRT family proteins are highly conserved NAD+ dependent class IIII HDACs.25 All seven known mammalian SIRTs harbor a conserved NAD+ binding domain and catalytic functional domain as well as distinctive N- and C-terminal domains which enable them to bind to a range of substrates.26 SIRT7 has been linked to important physiological processes such as rRNA transcription and modification, cell stress responses, DNA repair, and cellular metabolic activity.27,28 SIRT7 is closely linked to both aging and cardiac disease, and there is also evidence that it may shape tumorigenesis and associated pathological processes.
In one recent study, SIRT7 overexpression was shown to drive the growth of HCC tumors via the suppression of p21WAF1/Cip1 and the activation of cyclin D1.29 In gastric cancer, SIRT7 was instead shown to bind to the miR-34a promoter and to suppress its H3K18 acetylation, thus decreasing the expression of this miRNA, inhibiting apoptotic cell death, and thereby enhancing the growth of gastric tumors.30 By modulating CDC4 transcription, SIRT7 can suppress cytokinesis to promote the proliferation, migration, and invasion of osteosarcoma cells.31 In breast cancer, SIRT7 has also been shown to suppress SMAD4-dependent metastatic progression, suggesting that this may be a process that is amenable to therapeutic targeting.32 SIRT7 has also been suggested to promote the activation of the ERK/STAT3 signaling pathway in glioma cells, thereby favoring proliferation and invasion. Together, these prior reports suggest that SIRT7 can play distinct tumor-specific roles in many cancers.
As there have been relatively few studies to date exploring the expression of SIRT7 in KIRC, we herein conducted an integrated bioinformatics analysis of its potential diagnostic and functional relevance in this oncogenic setting. We found that KIRC tumors exhibited marked SIRT7 upregulation which was correlated with patient ethnicity, T stage, M stage, and pathologic stage. The overexpression of SIRT7 was associated with a poor patient prognosis and decreased OS, suggesting that SIRT7 may function in an oncogenic manner in the context of KIRC development. Increased SIRT7 expression was also negatively correlated with lung cancer patient OS in a prior study,33 and ROC analyses have demonstrated the value of SIRT7 as a prognostic biomarker in pancreatic cancer.34
To gain further insight into the functional role of SIRT7 as a driver of KIRC progression, we conducted pathway analyses which revealed this gene to be linked to both ubiquitin-mediated proteolysis and nucleotide excision repair. This gene may thus regulate protein homeostasis and DNA damage repair in KIRC cells. Recent findings suggest that glucose deprivation or chemotherapy hijacks the GSK3β-SIRT7 axis through ERK2, thereby activating AKT and affecting tumor malignant progression.35 SIRT7 deficiency in breast cancer cells after lncrna and mrna was mainly involved in regulating carboxylic acid metabolic processes and in the glycolytic pathway.36
The tumor microenvironment (TME) is composed of both tumor cells as well as a variety of immune cells and stromal cell types such as neurons, endothelial cells, and fibroblasts.37,38 Analyses of immune cell infiltration within the TME can aid in the prediction of patient immunotherapeutic responsiveness.39,40 As such, we explored the association between SIRT7 expression and tumor infiltration by different immune cell types in KIRC tumors, revealing SIRT7 expression to be moderately associated with infiltration by NK CD56bright cells, pDCs, macrophages, and Th2 cells. These analyses further suggested that SIRT7 may attenuate the functions of gd T cells, Mast cells, iDCs, Macrophages, Neutrophils, and Th2 cells in KIRC tumors. T cells are important group members in the tumor immune microenvironment, and T cells can be broadly divided into αβ T cells and γδ T cells. There is a significant negative correlation between γδ T cells and Treg cells in hepatocellular carcinoma, and Treg cells inhibit the killing function of γδ T cells through transforming growth factor β and interleukin 10-dependent pathways;41 in addition, animal experiments have shown that γδ T cells can inhibit tumorigenesis and progression by secreting IFN-γ and inhibiting the pro-oncogenic components in αβ T cells.42 Many studies have found that histone deacetylation family inhibitors can promote the expression of MHC in tumor cells and enable NK cells to recognize and kill tumor cells, a process that is mainly achieved through NKG2D.43 NKG2D is mainly expressed in natural killer cells, CD8+ T cells and γδ T cells and plays a role in tumor antigen recognition and tumor cell killing.44 Our results suggest that Sirt7 may affect the differentiation of T cells to some extent, participate in the impaired function of γδ T cells and NK cells in cancer tissues, and promote the growth of tumor cells, and the specific molecular mechanisms deserve further investigation. SIRT7 expression was also significantly correlated with the expression of many chemokines, chemokine receptors, and immune checkpoint genes in KIRC tumors, suggesting a range of mechanisms whereby it may shape the tumor immune microenvironment.
This study is the first to our knowledge to have explored the association between SIRT7 and KIRC patient outcomes. However, these results are subject to certain limitations. For one, these analyses were primarily based upon bioinformatics analyses, and direct experimental validation is thus warranted. Additionally, there were relatively differences in the number of healthy subjects (controls) and cancer patients. Furthermore, non-uniform interventions were employed in retrospective studies. Follow-up research will thus be essential to confirm the validity of our results.
Conclusions
In summary, the present study demonstrated that KIRC patients exhibit SIRT7 upregulation, which is correlated with a shorter survival duration. SIRT7 may additionally play a role in mediating the progression of KIRC by modulating the ubiquitin-mediated proteolysis and nucleotide excision repair pathways. Together, these data thus highlight SIRT7 as a promising diagnostic and prognostic biomarker in KIRC.
Data Sharing Statement
The data underlying this study are freely available from TCGA data portal (https://portal.gdc.cancer.gov/projects/TCGA-KIRC) and the GSE563757 and GSE40435 dataset (http://www.ncbi.nlm.nih.gov/geo/). The authors did not have special access privileges.
Ethics Statement
The Ethics Committee of the Guizhou Nursing Vocational College agreed to submit the study for review and has waived the need for ethical approval, as the data in this study are from public bioinformatics databases.
Author Contributions
All authors made substantial contributions to write and data visualization of the article, and revised the manuscript; revised the manuscript and data statistics; participated in data analysis and statistics of the data; assisted in literature search and data processing using software; checked and corrected the language expression and the layout of the manuscript. All authors read and approved the manuscript and agreed to take responsibility for all aspects of the study.
Funding
This work was supported by grants from National Natural Science Foundation of China (Nos.31771290) Guizhou Nursing Vocational College Foundation (gzhlyj2021-02), and Science and Technology Foundation of Guizhou Provincial Health Committee (gzwkj2022-518).
Disclosure
The authors declare that they have no competing interests.
References
1. Linehan WM, Ricketts CJ. The cancer genome atlas of renal cell carcinoma: findings and clinical implications. Nat Rev Urol. 2019;16(9):539–552. doi:10.1038/s41585-019-0211-5
2. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. doi:10.1038/nrdp.2017.9
3. Mun EJ, Babiker HM, Weinberg U, Kirson ED, Von Hoff DD. Tumor-treating fields: a fourth modality in cancer treatment. Clin Cancer Res. 2018;24(2):266–275. doi:10.1158/1078-0432.CCR-17-1117
4. Vanneman M, Dranoff G. Combining immunotherapy and targeted therapies in cancer treatment. Nat Rev Cancer. 2012;12(4):237–251. doi:10.1038/nrc3237
5. Yin L, Li W, Wang G, et al. NR1B2 suppress kidney renal clear cell carcinoma (KIRC) progression by regulation of LATS 1/2-YAP signaling. J Exp Clin Cancer Res. 2019;38(1):343. doi:10.1186/s13046-019-1344-3
6. Hu J, Chen Z, Bao L, et al. Single-cell transcriptome analysis reveals intratumoral heterogeneity in ccRCC, which results in different clinical outcomes. Mol Ther. 2020;28(7):1658–1672. doi:10.1016/j.ymthe.2020.04.023
7. Dai H, Sinclair DA, Ellis JL, Steegborn C. Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharmacol Ther. 2018;188:140–154. doi:10.1016/j.pharmthera.2018.03.004
8. Carafa V, Rotili D, Forgione M, et al. Sirtuin functions and modulation: from chemistry to the clinic. Clin Epigenetics. 2016;8:61. doi:10.1186/s13148-016-0224-3
9. Jiang Y, Liu J, Chen D, Yan L, Zheng W. Sirtuin inhibition: strategies, inhibitors, and therapeutic potential. Trends Pharmacol Sci. 2017;38(5):459–472. doi:10.1016/j.tips.2017.01.009
10. Mautone N, Zwergel C, Mai A, Rotili D. Sirtuin modulators: where are we now? A review of patents from 2015 to 2019. Expert Opin Ther Pat. 2020;30(6):389–407. doi:10.1080/13543776.2020.1749264
11. Bi S, Liu Z, Wu Z, et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell. 2020;11(7):483–504. doi:10.1007/s13238-020-00728-4
12. Yan WW, Liang YL, Zhang QX, et al. Arginine methylation of SIRT7 couples glucose sensing with mitochondria biogenesis. EMBO Rep. 2018;19(12):e46377. doi:10.15252/embr.201846377
13. Li H, Tian Z, Qu Y, et al. SIRT7 promotes thyroid tumorigenesis through phosphorylation and activation of Akt and p70S6K1 via DBC1/SIRT1 axis. Oncogene. 2019;38(3):345–359. doi:10.1038/s41388-018-0434-6
14. Yu H, Ye W, Wu J, et al. Overexpression of sirt7 exhibits oncogenic property and serves as a prognostic factor in colorectal cancer. Clin Cancer Res. 2014;20(13):3434–3445. doi:10.1158/1078-0432.CCR-13-2952
15. Zhao J, Wozniak A, Adams A, et al. SIRT7 regulates hepatocellular carcinoma response to therapy by altering the p53-dependent cell death pathway. J Exp Clin Cancer Res. 2019;38(1):252. doi:10.1186/s13046-019-1246-4
16. Ding M, Jiang CY, Zhang Y, Zhao J, Han BM, Xia SJ. SIRT7 depletion inhibits cell proliferation and androgen-induced autophagy by suppressing the AR signaling in prostate cancer. J Exp Clin Cancer Res. 2020;39(1):28. doi:10.1186/s13046-019-1516-1
17. Monteiro-Reis S, Lameirinhas A, Miranda-Gonçalves V, et al. Sirtuins’ deregulation in bladder cancer: SIRT7 is implicated in tumor progression through epithelial to mesenchymal transition promotion. Cancers. 2020;12(5):1066. doi:10.3390/cancers12051066
18. Vivian J, Rao AA, Nothaft FA, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35(4):314–316. doi:10.1038/nbt.3772
19. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi:10.1158/0008-5472.CAN-17-0307
20. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi:10.1093/nar/gkaa407
21. Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi:10.1016/j.immuni.2013.10.003
22. Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–461. doi:10.1016/j.ccell.2015.03.001
23. Akinleye A, Rasool Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J Hematol Oncol. 2019;12(1):92. doi:10.1186/s13045-019-0779-5
24. Pai CS, Simons DM, Lu X, et al. Tumor-conditional anti-CTLA4 uncouples antitumor efficacy from immunotherapy-related toxicity. J Clin Invest. 2019;129(1):349–363. doi:10.1172/JCI123391
25. O’Callaghan C, Vassilopoulos A. Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell. 2017;16(6):1208–1218. doi:10.1111/acel.12685
26. van de Ven RAH, Santos D, Haigis MC. Mitochondrial sirtuins and molecular mechanisms of aging. Trends Mol Med. 2017;23(4):320–331. doi:10.1016/j.molmed.2017.02.005
27. Wu D, Li Y, Zhu KS, Wang H, Zhu WG. Advances in cellular characterization of the sirtuin isoform, SIRT7. Front Endocrinol. 2018;9:652. doi:10.3389/fendo.2018.00652
28. Yamagata K, Yoshizawa T. Transcriptional regulation of metabolism by SIRT1 and SIRT7. Int Rev Cell Mol Biol. 2018;335:143–166. doi:10.1016/bs.ircmb.2017.07.009
29. Kim JK, Noh JH, Jung KH, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57(3):1055–1067. doi:10.1002/hep.26101
30. Yu W, Cui X, Wan Z, Yu Y, Liu X, Jin L. Silencing forkhead box M1 promotes apoptosis and autophagy through SIRT7/mTOR/IGF2 pathway in gastric cancer cells. J Cell Biochem. 2018;119(11):9090–9098. doi:10.1002/jcb.27168
31. Wei W, Jing ZX, Ke Z, Yi P. Sirtuin 7 plays an oncogenic role in human osteosarcoma via downregulating CDC4 expression. Am J Cancer Res. 2017;7(9):1788–1803.
32. Tang X, Shi L, Xie N, et al. SIRT7 antagonizes TGF-β signaling and inhibits breast cancer metastasis. Nat Commun. 2017;8(1):318. doi:10.1038/s41467-017-00396-9
33. Sun C, Zeng X, Guo H, et al. MicroRNA-125a-5p modulates radioresistance in LTEP-a2 non-small cell lung cancer cells by targeting SIRT7. Cancer Biomark. 2020;27(1):39–49. doi:10.3233/CBM-190381
34. McGlynn LM, McCluney S, Jamieson NB, et al. SIRT3 & SIRT7: potential novel biomarkers for determining outcome in pancreatic cancer patients. PLoS One. 2015;10(6):e0131344. doi:10.1371/journal.pone.0131344
35. Tang X, Li G, Shi L, et al. Combined intermittent fasting and ERK inhibition enhance the anti-tumor effects of chemotherapy via the GSK3β-SIRT7 axis. Nat Commun. 2021;12(1):5058. doi:10.1038/s41467-021-25274-3
36. Chen KL, Li L, Wang YR, Li CM, Badri TM, Wang GL. Long noncoding RNA and mRNA profiling in MDA-MB-231 cells following RNAi-mediated knockdown of SIRT7. Onco Targets Ther. 2017;10:5115–5128. doi:10.2147/OTT.S149048
37. Hessmann E, Buchholz SM, Demir IE, et al. Microenvironmental determinants of pancreatic cancer. Physiol Rev. 2020;100(4):1707–1751. doi:10.1152/physrev.00042.2019
38. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi:10.1038/s41591-018-0014-x
39. Belli C, Trapani D, Viale G, et al. Targeting the microenvironment in solid tumors. Cancer Treat Rev. 2018;65:22–32. doi:10.1016/j.ctrv.2018.02.004
40. Zhang W, Qian Y, Jin X, et al. SIRT7 is a prognostic biomarker in kidney renal clear cell carcinoma that is correlated with immune cell infiltration. PREPRINT (Version 2) available at Research Square; 2021. 10.21203/rs.3.rs-867349/v2.
41. Yi Y, He HW, Wang JX, et al. The functional impairment of HCC-infiltrating γδ T cells, partially mediated by regulatory T cells in a TGFβ- and IL-10-dependent manner. J Hepatol. 2013;58(5):977–983. doi:10.1016/j.jhep.2012.12.015
42. Girardi M. Immunosurveillance and immunoregulation by gammadelta T cells. J Invest Dermatol. 2006;126(1):25–31. doi:10.1038/sj.jid.5700003
43. Skov S, Pedersen MT, Andresen L, Straten PT, Woetmann A, Odum N. Cancer cells become susceptible to natural killer cell killing after exposure to histone deacetylase inhibitors due to glycogen synthase kinase-3-dependent expression of MHC class I-related chain A and B. Cancer Res. 2005;65(23):11136–11145. doi:10.1158/0008-5472.CAN-05-0599
44. Bauer S, Groh V, Wu J, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285(5428):727–729. doi:10.1126/science.285.5428.727
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.