Back to Journals » OncoTargets and Therapy » Volume 14
Sinomenine Inhibits the Growth of Ovarian Cancer Cells Through the Suppression of Mitosis by Down-Regulating the Expression and the Activity of CDK1
Authors Qu X, Yu B , Zhu M , Li X, Ma L, Liu C, Zhang Y, Cheng Z
Received 28 September 2020
Accepted for publication 8 December 2020
Published 5 February 2021 Volume 2021:14 Pages 823—834
DOI https://doi.org/10.2147/OTT.S284261
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr XuYu Yang
Xiaoyan Qu,1,* Bing Yu,2,* Mengmei Zhu,2,* Xiaomei Li,2,3 Lishan Ma,1 Chuyin Liu,1 Yixing Zhang,1 Zhongping Cheng4
1Department of Gynecology and Obstetrics, Yangpu Hospital, Tongji University School of Medicine, Shanghai, 200090, People’s Republic of China; 2Department of Cell Biology, Navy Medical University (Second Military Medical University), Shanghai, 200433, People’s Republic of China; 3Cancer Research Laboratory, The Affiliated Hospital of Zunyi Medical University, Zunyi, Guizhou Province, 563003, People’s Republic of China; 4Department of Gynecology and Obstetrics, Shanghai Tenth People’s Hospital, Tongji University School of Medicine, Shanghai, 200072, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhongping Cheng
Department of Gynecology and Obstetrics, Shanghai Tenth People’s Hospital, Tongji University School of Medicine, 301 Yanchang Middle Road, Shanghai, 200072, People’s Republic of China
Tel +86-021-66300588
Email [email protected]
Introduction: Ovarian cancer is one of the most common gynecological cancers worldwide. While, therapies against ovarian cancer have not been completely effective, sinomenine has been proved to have anti-tumor activity in various cancer cells. However, study of its anti-ovarian cancer effect is still rare, and the underlying mechanism has not been elucidated. Therefore, we aim to explore the mechanism of sinomenine anti-ovarian cancer.
Materials and Methods: The effect of anti-ovarian cancer HeyA8 cells was analyzed by CCK8 and colony formation assay. The mechanism of sinomenine anti-ovarian cancer was explored via high throughput RNA-seq, and then the target mRNA and protein expression were verified by real-time PCR and Western blot, respectively.
Results: We found that the proliferation and clone formation ability of ovarian cancer HeyA8 cells were markedly reduced by 1.56 mM sinomenine. The transcriptome analysis showed that 2679 genes were differentially expressed after sinomenine treatment in HeyA8 cells, including 1323 down-regulated genes and 1356 up-regulated genes. Gene ontology and KEGG pathway enrichment indicated that differential expression genes (DEGs) between the groups of sinomenine and DMSO-treated HeyA8 cells were mainly involved in the process of the cell cycle, such as kinetochore organization, chromosome segregation, and DNA replication. Strikingly, the top 18 ranked degree genes in the protein–protein interaction (PPI) network were mainly involved in the process of mitosis, such as sister chromatid segregation, condensed chromosome, and microtubule cytoskeleton organization. Moreover, real-time PCR results showed consistent expression trends of DEGs with transcriptome analysis. The results of Western blot showed the expression level of CDK1, which was the highest degree gene in PPI and the main regulator controlling the process of mitosis, and the levels of phosphorylated P-CDK (Thr161) and P-Histone H3 (Ser10) were decreased after being treated with sinomenine.
Conclusion: Our results demonstrated that sinomenine inhibited the proliferation of HeyA8 cells through suppressing mitosis by down-regulating the expression and the activity of CDK1. The study may provide a preliminary research basis for the application of sinomenine in anti-ovarian cancer.
Keywords: ovarian cancer, sinomenine, high throughput RNA-seq, CDK1, cell proliferation
Introduction
Ovarian cancer (OC) is the seventh most common cancer among women in the world and the second leading cause of gynecological cancer death.1 Most OC is often diagnosed at the advanced stage.2 In the past few decades, many treatment methods, such as active surgery, targeted therapy, intraperitoneal hyperthermia chemotherapy, small molecular inhibitors, neoadjuvant chemotherapy, and intraperitoneal chemotherapy, were hired to cure OC.3,4 However, most patients with ovarian cancer will suffer from tumor recurrence after first-line therapy. Although OC is sensitive to chemotherapy with platinum and taxane following debulking surgery, resistance to chemotherapy will eventually develop in almost all patients. Once the disease recurs, the interval between subsequent treatments steadily decreases due to rapid development and chemoresistance.5 Therefore, it is urgent to explore more effective reagents for the treatment of OC.
Natural plant products have been widely used in the treatment of various diseases. Sinomenine (7,8-didehydro-4-hydroxy-3,7-dimethoxy-17-methylmorphinan-6-one), an alkaloid monomer extracted from Sinomenium acutum plants, contains four rings, A, B, C and D.6 Many studies have proved that sinomenine has anti-inflammatory, anti-rheumatic, anti-oxidant, analgesic, immunosuppressive, and anti-angiogenic effects.7–11 Although the shorter biological half-life of sinomenine and the side-effects, such as increasing histamine release, restrained its clinical applications,6,12 sinomenine has also been developed into Zhengqingfengtongning (ZQFTN), a Chinese proprietary medicine approved by the Chinese government for treating RA and other autoimmune diseases in China. The anti-tumor effect of sinomenine has been preliminarily addressed in many kinds of tumors, such as liver cancer,13 breast cancer,14 lung cancer,15 renal cell carcinoma,16 and glioblastoma.17 Recently, the inhibitory effect of sinomenine on growth and metastasis of ovarian cancer cells has drawn considerable attention.18,19 Li et al showed that sinomenine inhibits ovarian cancer cell growth and metastasis by inhibiting the Wnt/β-catenin pathway via targeting MCM2.18 Xu et al identified that sinomenine exerted the antitumor effect in ovarian cancer cells by hindering the expression of long non-coding RNA HOST2.19 However, these studies and findings are basic and preliminary for the effect of sinomenine on ovarian cancer. The mechanism of sinomenine inhibiting ovarian cancer remains to be further elucidated.
High-throughput sequencing, especially RNA sequencing (RNA-seq) has become an effective way to explore functional genes and mechanisms in anti-tumor research because of its low cost and ultra-high data output.20 Recently, RNA-seq has been successfully applied in ovarian cancer research for earlier detection, identification of pathological origin, defining the aberrant genes and dysregulated molecular pathways across patient groups, and identification of novel genes and molecular pathways in the development of multidrug resistance.21 Here, we hired high-throughput RNA-seq to explore the potential mechanism of the sinomenine mediated growth inhibition of ovarian cancer HeyA8 cells. Then, the results of the high-throughput mRNA sequence were validated by real-time PCR. Furthermore, we also preliminarily clarified that sinomenine inhibited the growth of ovarian cancer cells through the suppression of mitosis by down-regulating the expression and the activity of CDK1. This study was expected to lay a theoretical foundation for the future application of sinomenine in the treatment of ovarian cancers.
Materials and Methods
Tumor Cells and Cell Culture Condition
The human ovarian cancer cell line HeyA8-MDR was obtained from Stem Cell Bank, Chinese Academy of Sciences and was maintained in RPMI-1640 (Hyclone) supplemented with 10% fetal bovine serum (Gibcol) and 1% penicillin/streptomycin (Gibcol) at 37 °C in a humidified atmosphere with 5% CO2.
Cell Survival Rate Assay and Cell Proliferation Assay
For cell survival rate assay, HeyA8-MDR cells were seeded at a density of 5000 cells per well into 96-well plates. After incubation for 24 hours, sinomenine (0, 0.25, 0.5, 1, 2, 4 and 8 mM) was added to each well. The control cells (0 µM) were only treated with an equivalent volume of DMSO. Each concentration of sinomenine was tested with six replicates. After incubation for 48 hours, the culture medium was supplemented with 10 ul CCK8 solution for 2 hours at 37 °C. Then the absorbance was measured at 450 nm. Cell survival rates were shown as a percentage of the absorbance reading of the control cells. IC50 was calculated with the IC50 Calculator (https://www.aatbio.com/tools/ic50-calculator).
Cell proliferation assay was detected by CCK8 as described in our previous study.22 Briefly, 1 × 103 HeyA8-MDR cells per well were plated on 96-well plates. After being incubated for 24 hours, the cells were treated with 1.56 mM sinomenine and DMSO separately. Then, 10 µL CCK8 reagent was added to each well and incubated for 2 hours at 37 °C. The optical density value was measured at 450 nm every day for 7 days.
Clone Formation Assay
HeyA8-MDR cells were seeded at a density of 200 cells per well into 6-well plates. HeyA8-MDR cells, treated with 1.56 mM sinomenine and DMSO separately, were incubated for 10 days in a cell incubator at 37 °C. Then, the cells were fixed with 4% PFA and stained with crystal violet staining solution (#Y1232, Yuxiu Biotech, China). The number of colonies, containing more than 50 cells observed under a microscope, were counted.
RNA Extraction, RNA-Seq Transcriptome Library Construction, and Illumina HiSeq 4000 Sequencing
RNA extraction, library construction and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China). Total RNA was extracted using TRIzol® Reagent (Thermo Scientific) and genomic DNA was removed by digestion with DNase I (Takara). Then RNA quality was determined by 2100 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). Only high-quality RNA samples (OD260/280 = 1.8~2.2, OD260/230≥2.0, RIN≥6.5, 28S:18S≥1.0, >2 μg) were used to construct the sequencing library.
RNA-seq transcriptome library was constructed with the TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA) using 1 μg of total RNA. Shortly, messenger RNA was purified using oligo (dT) magnetic beads and then fragmented by fragmentation buffer. Double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen) with random hexamer primers (Illumina). Then the synthesized cDNA was subjected to end-repair, phosphorylation and “A” base addition according to Illumina’s library construction protocol. Libraries were size selected for cDNA target fragments of 200–300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After being quantified by TBS380, the paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq 4000 (2×150 bp read length). The RNA-seq data were deposited at NCBI (BioProject id: PRJNA641485).
Read Mapping
The raw paired end reads were trimmed and quality controlled by SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) with default parameters. Then clean reads were separately aligned to reference the genome with orientation mode using TopHat (http://tophat.cbcb.umd.edu/, version2.1.1) software.23 The mapping criteria of bowtie were as follows: sequencing reads should be uniquely matched to the genome allowing up to 2 mismatches, without insertions or deletions. Then the region of the gene was expanded following depths of sites and the operon was obtained. In addition, the whole genome was split into multiple 15 kb windows that share 5 kb. New transcribed regions were defined as more than 2 consecutive windows without an overlapped region of gene, where at least 2 reads were mapped per window in the same orientation.
Differential Expression Analysis and Functional Enrichment
The expression level of each transcript was calculated according to the Transcripts Per Kilobase of exon model per Million mapped reads (TPM) using RSEM (http://deweylab.biostat.wisc.edu/rsem/).24 The differentially expressed genes (DEGs) (fold changes ≥ 2 and corrected P-value ≤0.05) between control and sinomenine-treated HeyA8 cell line were identified by R statistical package software EdgeR (Empirical analysis of Digital Gene Expression in R, http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html).25 GO functional enrichment and KEGG pathway analysis were carried out by Goatools (https://github.com/tanghaibao/Goatools) and KOBAS 2.1.1 (http://kobas.cbi.pku.edu.cn/download.php).26 The GO terms and the KEGG pathways were considered statistically significant when Bonferroni-corrected P-value ≤0.05.
Protein–Protein Interaction (PPI) Network Analysis
The PPI for DEGs (fold changes≥4 and corrected P-value ≤0.05) were calculated on the web of the Retrieval of Interacting Genes (STRING) database (https://string-db.org/)27 and the cut-off criterion of the combined score was set as >0.4. Then, PPI networks for DEGs were visualized by Cytoscape (Version: 3.8.0).28
Real-Time PCR
Total RNAs were reverse transcribed by the MMLV Reverse Transcriptase (Promega) according to the manufacture’s protocol. Real-time PCR analysis was performed on the LightCycler® 96 System (Roche) with ChamQ SYBR qPCR Master Mix (Q321-02, Vazyme, China). All samples were examined in triplicate. The fold changes of each target gene were calculated using the 2−ΔΔCt method relative to GAPDH.
Flow Cytometry Assay
Cells, which were treated with sinomenine and DMSO separately, were harvested and washed twice with PBS. Then the cells were fixed with 4% paraformaldehyde for 10 min at 4 °C, and permeabilized for 10 minutes with 0.3% Triton X-100. The cells were incubated with anti-P-Histone H3 (53348, CST) antibodies for 30 minutes. After staining, the cells were washed twice and then incubated with FITC-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) (SA00003-2, Proteintech) for 30 minutes. After being washed twice, the stained cells were stained with 50 ug/mL propidium iodide (PI) and analyzed with Beckman Coulter CytoFLEX flow cytometry system.
Western Blot
Western blots were performed as described previously.29 The primary antibodies were listed as follows: Geminin (52508), CDT1 (8064), Thymidine Kinase 1 (28755), P-Histone H3 (53348), Cyclin A2 (91500), Cyclin B1 (12231), Cyclin E1 (20808) and P-cdc2 (Tyr15) (4539) from Cell Signaling Technology, P-Cdk1/2 (Thr14) (DF2944) and P-CDK1 (Thr161) (AF8001) from Affinity, CDK1 (67575-1-Ig) from Protein Tech Group.
Statistical Analysis
The data were shown as means ± standard deviation (SD). The difference between means was analyzed using SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). The methods of statistical analysis were indicated in figure legends. p < 0.05 was considered as statistically significant.
Results
Sinomenine Inhibits the Growth of Ovarian Cancer Cell Line HeyA8
To explore the inhibitory effect of sinomenine on the cell growth of ovarian cancer HeyA8 cells, the cell viability of HeyA8 were measured by CCK8 assay. As shown in Figure 1A, the inhibitory effect of sinomenine on HeyA8 cells was dose-dependent. The IC50 of sinomenine was 1.56 mM in HeyA8 cells (Figure 1B). The result of the CCK8 assay showed that sinomenine significantly suppressed the proliferation of HeyA8 at the concentration of 1.56 mM (Figure 1C). In addition, the result of the clone formation assay also indicated that sinomenine significantly inhibit the ability of the clone formation of HeyA8 cells (Figure 1D and E). Taken together, these results indicated that sinomenine inhibited the growth of ovarian cancer HeyA8 cells, which is consistent with results in previous reports.18,19
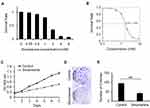 |
Figure 1 Sinomenine inhibits proliferation of ovarian cancer cell line HeyA8. (A) The survival rate of HeyA8 cells treated with different concentrations of sinomenine (0, 0.25, 0.5, 1.0, 2.0, 4.0, and 8.0 mM) for 48 hours were measured by CCK8 assay. (B) The IC50 of sinomenine was 1.56 mM in HeyA8 cells, which was calculated with the IC50 Calculator on the website (https://www.aatbio.com/tools/ic50-calculator). (C) The growth curves of HeyA8 treated with DMSO as control and sinomenine (1.56 mM) were measured by CCK8 assay. (D) HeyA8 treated with sinomenine formed fewer and smaller colonies than those treated with DMSO. (E) The number of colonies of HeyA8 cells treated with sinomenine and DMSO separately in (D), the value represents the mean ± SD for triplicate samples, **P<0.01, Student’s t-test. |
Differentially Expressed Genes (DEGs) in HeyA8 Cells Caused by the Treatment of Sinomenine
To investigate the DEGs caused by the treatment of sinomenine, the gene expression of HeyA8 cells, which were treated with 1.56 mM sinomenine and an equal volume DMSO for 48 hours separately, were analyzed by high-throughput RNA sequencing. After gene mapping and the expression level of each transcript was converted to the value of TPM, the comparison at corrected P-value p ≤ 0.05 and log2FC fold change ≥ 1 (for up-regulation) or ≤ −1 (for down-regulation) was made to identify the DEGs for two groups (Figure 2A). The list of DEGs, along with their TPM and annotations, are presented in Supplementary Table S1. A total of 2679 genes were identified as DEGs, including 1323 down-regulated genes and 1356 up-regulated genes (Figure 2B), and were displayed by cluster heatmaps (Figure 2C).
Gene Ontology and KEGG Pathway Enrichment Analysis of DEGs
To study the characteristics of the 2679 DEGs, gene ontology (GO) enrichment analysis was performed. The top 20 ranked GO terms of DEGs were shown in Figure 3A. Kinetochore organization occupied the strongest enrichment degree as it possessed the highest rich factor (0.65), followed by strand displacement, meiotic chromosome segregation, attachment of spindle microtubules to kinetochore and DNA strand elongation. These enriched GO terms were obviously involved in the regulation of cell cycle, especially involved in the DNA replication in S phase, Kinetochore organization in G2 phase and chromosome segregation in M phase. These indicated that sinomenine inhibits the HeyA8 cell growth by regulating the process of the cell cycle.
To further investigate the biological functions of DEGs, KEGG pathway enrichment analysis was performed. The results also showed the top 20 statistically significant pathways (Figure 3B). The top 5 significant pathways are DNA replication, cell cycle, homologous recombination, Fanconi anemia pathway, and microRNAs in cancer. In addition to the above pathways, these pathways including purine metabolism, pyrimidine metabolism, mismatch repair and nucleotide excision repair are also closely related to the cell cycle. Taken together, these results of GO and KEGG pathway enrichment could provide essential information on the investigation of sinomenine in ovarian cancer HeyA8 cells.
Protein–Protein Interaction (PPI) Network Analysis of DEGs
In order to systemically analyze the functions of DEGs in sinomenine-treated ovarian cancer HeyA8 cells, a total of 856 DEGs (foldchange ≥ 4, corrected P-value ≤ 0.05) were mapped to the PPI database to obtain the PPI networks. As shown in Figure 4A, a total of 600 relationships between 104 genes (nodes) were identified. Table 1 shows the node genes with the top 18 ranked degree (number of interactions). The DEGs of CDK1 (degree = 62), PLK1 (degree = 50), BUB1 (degree = 48), NDC80 (degree = 48) and BUB1B (degree = 44) formed networks with high degrees. The top 3 ranked degree genes (CDK1, PLK1, and BUB1) play important roles in the progression of G2/M phase in the cell cycle.30,31 Furthermore, real-time PCR analysis was used to verify the results of transcriptome sequencing and the results indicated that the expression trends of the top 18 ranked degree genes were consistent with those obtained by RNA-seq, suggesting that the RNA-seq data reliably reflected the gene expression alterations (Figure 4B). According to the RNA-seq and real-time PCR results, the expressions of cell cycle regulated genes, such as CDK1, PLK1, and BUB1, were significantly decreased after sinomenine treatment. These results were also in accord with the cell phenotype experiments.
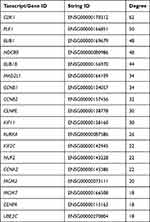 |
Table 1 Node Genes with Top 18 Ranked Degree in the PPI Network |
In addition, GO enrichment analysis indicated that the validated 18 genes were enriched in regulation of sister chromatid segregation, chromosome, condensed chromosome, microtubule cytoskeleton organization involved in mitosis, condensed nuclear chromosome, mitotic cell cycle process, and mitotic nuclear division, etc. (Figure 4C). These results suggested that sinomenine may affect the proliferation of ovarian cancer HeyA8 cells by regulating the progression of M phase in cell cycle through these target genes and their classical pathways.
Sinomenine Suppressed Mitosis by Inhibiting the Expression and the Activity of CDK1
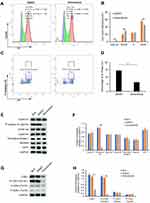
The results of RNA-seq and real-time PCR indicated that sinomenine inhibited the mRNA expression level of CDK1 in ovarian cancer HeyA8 cells. While, the cyclin B1/Cdk1 complex was reported to play an important role in regulating the entry into mitosis.32,33 In order to clarify the effect of sinomenine on cell cycle distribution, firstly, we analyzed the cell cycle distribution of HeyA8 cells by flow cytometry assay and the results showed the population of G2/M were decreased from 55.6% ± 2.85% to 44.08% ± 3.22% in HeyA8 cells treated with sinomenine for 48 hours (Figure 5A and B). Meanwhile, the percentage of M phase was decreased from 14.46 ± 1.10% to 6.35% ± 0.61% (Figure 5C and D). Then, the expression levels of some cell cycle related proteins including cyclin E1, cyclin A2, cyclin B1, Geminin, CDT1, Thymidine kinase 1 and P-Histone H3 (Ser10), which presented in cells at various phases of the cell cycle, were analyzed by Western blot (Figure 5E). The results showed that the expression level of P-Histone H3 (Ser10), which is present only in the M phase, was significantly decreased in sinomenine treated HeyA8 cells when compared with DMSO treated negative control HeyA8 cells and blank control HeyA8 cells. However, there were no significant changes in the expression levels of other cell cycle related proteins (Figure 5F). These results indicated that sinomenine decreased the number of M-phase cells, suggesting the suppression of mitosis in ovarian cancer HeyA8 cells.
Meanwhile, the activity of CDK1 depends on its phosphorylation state.34 Phosphorylation of the Thr161 residue of Cdk1 can stabilize its interaction with cyclins and lead to further activation of the kinase.35 On the other hand, the cyclin B1/Cdk1 complex can be inactivated via inhibitory phosphorylation of Thr14 and Tyr15 residues of the CDK1.36 Therefore, we detected the protein expression level and phosphorylation levels of CDK1 by Western blot (Figure 5G). The results showed that the expression level of CDK1 and the phosphorylation level of CDK1 on Thr161 in sinomenine treated HeyA8 cells were significantly lower than those in DMSO treated HeyA8 cells and control HeyA8 cells. While, the phosphorylation levels of CDK1 on Thr14 and Tyr15 did not change significantly (Figure 5H). These results indicated that sinomenine suppressed the expression of CDK1 and the activity of CDK1 by inhibiting the phosphorylation of CDK1 on Thr161.
Discussions
OC is the most lethal gynecologic malignancy and no significant treatment progress has been made in the past 30 years. It is easy to relapse and form drug resistance after the first comprehensive treatment. In order to improve the survival rate and prolong the survival period of patients, it is very important to find new effective chemotherapy drugs for the treatment of OC. Many studies showed that a large number of natural plant products possess the ability of anti-tumor. Up to now, many natural anti-tumor products, such as vinblastine, vincristine, podophyllotoxin, paclitaxel (Taxol) and camptothecin, have been tested clinically.37 In recent years, sinomenine, an alkaloid extracted from Sinomenium acutum plants, have been proved to have the ability of anti-tumor. However, the study of its anti-ovarian cancer effect is still rare and its mechanism is unclear. Here, we studied the anti-tumor effect of sinomenine on ovarian cancer cells and found that sinomenine inhibits the capacities of proliferation and colony formation of ovarian cancer HeyA8 cells, which was consistent with previous reports.18,19 Furthermore, we firstly preliminarily explored its mechanism by High-throughput RNA-seq. The results of GO and KEGG pathway enrichment and the Protein–Protein Interaction Analysis indicated that the DEGs were mainly involved in the cell cycle.
Cell survival and proliferation mainly depend on the progression of the cell cycle, which is a series of tightly-regulated molecular events controlling DNA replication and mitosis.38,39 The entry into mitosis is regulated by the activation of Cdc2/Cdk1 kinase, which was controlled by several steps including cyclin B1 nuclear accumulation and binding, dephosphorylation of Cdc2/Cdk1 at Thr14 and Tyr15, and phosphorylation of Cdc2/Cdk1 at Thr161.40,41 In our experiment, sinomenine decreased the expression level of CDK1 and especially suppressed the activated phosphorylation Thr161 on CDK1. At the same time, sinomenine also decreased the phosphorylation of Histone H3 at Ser10, which is tightly correlated with chromosome condensation during mitosis.42 These results suggested that sinomenine inhibited the proliferation of ovarian cancer HeyA8 cells through suppressing the mitosis by down-regulating the expression and the activity of CDK1.
While the process of the M phase is composed of many biological events, such as chromatin condenses, Spindle formation, chromosome separation, mitotic nuclear division, and mitotic DNA damage checkpoint.43 The results of GO enrichment assay indicated that many of the top 18 ranked DEGs in PPI were involved in multiple biological events in phase M. For instance, PLK1, serine/threonine-protein kinase 1, performs several important functions throughout M phase of the cell cycle, including the regulation of centrosome maturation and spindle assembly, the removal of cohesions from chromosome arms, the inactivation of anaphase-promoting complex/cyclosome (APC/C) inhibitors, and the regulation of mitotic exit and cytokinesis.44,45 Mitotic checkpoint serine/threonine-protein kinase BUB1 performs two crucial functions, including spindle-assembly checkpoint signaling and correct chromosome alignment, during mitosis.46 NDC80, as a component of the essential kinetochore-associated NDC80 complex, is required for chromosome segregation and spindle checkpoint activity.47 However, how sinomenine affects the process of M phase still needs to be further explored.
In summary, our results indicated that sinomenine plays anti-tumor functions by inhibiting the growth of the ovarian cancer HeyA8 cells through the suppression of mitosis by down-regulating the expression and the activity of CDK1. These results may provide a preliminary research basis for the application of sinomenine in anti-ovarian cancer.
Acknowledgments
This work was supported by the National Key R&D Program of China (2018YFA0107500), the Key Specialty Construction Project of the Shanghai Municipal Commission of Health and Family Planning (No. ZK2015A33, 201740117), National Natural Science Foundation of China (31771511) and Foundation strengthening program in technical field of China (2019-JCJQ-JJ-068).
Disclosure
The authors declare no conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
2. Tew WP. Ovarian cancer in the older woman. J Geriatr Oncol. 2016;7:354–361.
3. Pujade-Lauraine E. New treatments in ovarian cancer. Ann Oncol. 2017;28:viii57–viii60.
4. Cortez AJ, Tudrej P, Kujawa KA, et al. Advances in ovarian cancer therapy. Cancer Chemother Pharmacol. 2018;81:17–38.
5. Leung CS, Yeung TL, Yip KP, et al. Calcium-dependent FAK/CREB/TNNC1 signalling mediates the effect of stromal MFAP5 on ovarian cancer metastatic potential. Nat Commun. 2014;5:5092.
6. Zhang YS, Han JY, Iqbal O, et al. Research advances and prospects on mechanism of sinomenin on histamine release and the binding to histamine receptors. Int J Mol Sci. 2018;20:70.
7. Zhou H, Liu JX, Luo JF, et al. Suppressing mPGES-1 expression by sinomenine ameliorates inflammation and arthritis. Biochem Pharmacol. 2017;142:133–144.
8. Wu Y, Lin Z, Yan Z, et al. Sinomenine contributes to the inhibition of the inflammatory response and the improvement of osteoarthritis in mouse-cartilage cells by acting on the Nrf2/HO-1 and NF-κB signaling pathways. Int Immunopharmacol. 2019;75:105715.
9. Liu S, Chen Q, Liu J, et al. Sinomenine protects against E.coli-induced acute lung injury in mice through Nrf2-NF-κB pathway. Biomed Pharmacother. 2018;107:
10. Zhu Q, Sun Y, Zhu J, et al. Antinociceptive effects of sinomenine in a rat model of neuropathic pain. Sci Rep. 2014;4:7270.
11. Feng ZT, Yang T, Hou XQ, et al. Sinomenine mitigates collagen-induced arthritis mice by inhibiting angiogenesis. Biomed Pharmacother. 2019;13:108759.
12. Tang J, Raza A, Chen J, et al. A systematic review on the sinomenine derivatives. Mini Rev Med Chem. 2018;18:
13. Hong Y, Yang J, Shen X, et al. Sinomenine hydrochloride enhancement of the inhibitory effects of anti-transferrin receptor antibody-dependent on the COX-2 pathway in human hepatoma cells. Cancer Immunol Immunother. 2013;62:447–454.
14. Li X, Wang K, Ren Y, et al. MAPK signaling mediates sinomenine hydrochloride-induced human breast cancer cell death via both reactive oxygen species-dependent and -independent pathways: an in vitro and in vivo study. Cell Death Dis. 2014;5:e1356.
15. Shen KH, Hung JH, Liao YC, et al. Sinomenine inhibits migration and invasion of human lung cancer cell through downregulating expression of miR-21 and MMPs. Int J Mol Sci. 2020;21:3080.
16. Zhao B, Liu L, Mao J, et al. Sinomenine hydrochloride attenuates the proliferation, migration, invasiveness, angiogenesis and epithelial-mesenchymal transition of clear-cell renal cell carcinoma cells via targeting Smad in vitro. Biomed Pharmacother. 2017;96:1036–1044.
17. Jiang Y, Jiao Y, Liu Y, et al. Sinomenine hydrochloride inhibits the metastasis of human glioblastoma cells by suppressing the expression of matrix metalloproteinase-2/-9 and reversing the endogenous and exogenous epithelial-mesenchymal transition. Int J Mol Sci. 2018;19:844.
18. Li H, Lin Z, Bai Y, et al. Sinomenine inhibits ovarian cancer cell growth and metastasis by mediating the Wnt/β-catenin pathway via targeting MCM2. RSC Adv. 2017;7:50017–50026.
19. Xu Y, Jiang T, Wang C, et al. Sinomenine hydrochloride exerts antitumor outcome in ovarian cancer cells by inhibition of long non-coding RNA HOST2 expression. Artif Cells Nanomed Biotechnol. 2019;47:4131–4138.
20. Tang F, Barbacioru C, Wang Y, et al. mRNA-seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382.
21. Wang J, Dean DC, Hornicek FJ, et al. RNA sequencing (RNA-Seq) and its application in ovarian cancer. Gynecol Oncol. 2019;152:194–201.
22. Yu B, Li H, Chen J, et al. Extensively expanded murine-induced hepatic stem cells maintain high-efficient hepatic differentiation potential for repopulation of injured livers. Liver Int. 2020. doi:10.1111/liv.14509
23. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111.
24. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323.
25. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140.
26. Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322.
27. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368.
28. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504.
29. Yang LH, Wang Y, Qiao S, et al. Liver-enriched activator protein 1 as an isoform of ccaat/enhancer-binding protein beta suppresses stem cell features of hepatocellular carcinoma. Cancer Manag Res. 2018;10:873–885.
30. Liao H, Ji F, Ying S. CDK1: beyond cell cycle regulation. Aging. 2017;9:2465–2466.
31. Qi W, Tang Z, Yu H. Phosphorylation- and polo-box-dependent binding of Plk1 to bub1 is required for the kinetochore localization of Plk1. Mol Biol Cell. 2006;17:3705–3716.
32. Hunt T. Under arrest in the cell cycle. Nature. 1989;342:483–484.
33. Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508.
34. Woodbury EL, Morgan DO. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol. 2007;9:106–112.
35. Ducommun B, Brambilla P, Félix MA, et al. cdc2 phosphorylation is required for its interaction with cyclin. EMBO J. 1991;10:3311–3319.
36. Atherton-Fessler S, Liu F, Gabrielli B, et al. Cell cycle regulation of the p34cdc2 inhibitory kinases. Mol Biol Cell. 1994;5:989–1001.
37. Gezici S, Şekeroğlu N. Current perspectives in the application of medicinal plants against cancer: novel therapeutic agents. Anticancer Agents Med Chem. 2019;19:101–111.
38. Hall PA. Cell proliferation. J Pathol. 1991;165:349–354.
39. Matson JP, Cook JG. Cell cycle proliferation decisions: the impact of single cell analyses. FEBS J. 2017;284:362–375.
40. Mochida S, Rata S, Hino H, et al. Two bistable switches govern M phase entry. Curr Biol. 2016;26:3361–3367.
41. Norbury C, Blow J, Nurse P. Regulatory phosphorylation of the p34cdc2 protein kinase in vertebrates. EMBO J. 1991;10:3321–3329.
42. Hendzel MJ, Wei Y, Mancini MA, et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360.
43. McIntosh JR. Mitosis. Cold Spring Harb Perspect Biol. 2016;8:a023218.
44. Petronczki M, Lénárt P, Peters JM. Polo on the rise-from mitotic entry to cytokinesis with Plk1. Dev Cell. 2008;14:646–659.
45. Kishi K, van Vugt MA, Okamoto K, et al. Functional dynamics of Polo-like kinase 1 at the centrosome. Mol Cell Biol. 2009;29:3134–3150.
46. Raaijmakers JA, van Heesbeen R, Blomen VA, et al. BUB1 is essential for the viability of human cells in which the spindle assembly checkpoint is compromised. Cell Rep. 2018;22:1424–1438.
47. Amin MA, McKenney RJ, Varma D. Antagonism between the dynein and Ndc80 complexes at kinetochores controls the stability of kinetochore-microtubule attachments during mitosis. J Biol Chem. 2018;293:5755–5765.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.