Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 13
Single Nucleotide Variants Associated with Colorectal Cancer Among Iranian Patients: A Narrative Review
Authors Jamshidi M, Mohammadi Pour S, Mahmoudian-Sani MR
Received 4 February 2020
Accepted for publication 19 May 2020
Published 3 June 2020 Volume 2020:13 Pages 167—180
DOI https://doi.org/10.2147/PGPM.S248349
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Mohammad Jamshidi,1 Somayeh Mohammadi Pour,2 Mohammad-Reza Mahmoudian-Sani3
1Department of Laboratory Sciences, School of Allied Medicine, Lorestan University of Medical Sciences, Khorramabad, Iran; 2Department of Obstetrics and Gynecology, School of Medicine Lorestan University of Medical Sciences, Khorramabad, Iran; 3Thalassemia and Hemoglobinopathy Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
Correspondence: Mohammad-Reza Mahmoudian-Sani Tel +061-33750410
Fax +061-33750427
Email [email protected]
Abstract: Colorectal cancer has been considered as one of the complicated multi-stage processes after adenoma-carcinoma sequence. Therefore, studies of the molecular dysregulation basis could present information on the recognition of the potent biomarkers and treatment targets for this disease. Even though outcomes of the patients with colorectal cancer have been improved largely with current annual screening plans, it is necessary to have reliable prognostic biomarkers because of the disease heterogeneity. There is a significant relationship between SNP in IL1RN* 2 (IL1ra), − 509 C/T (TGFB1), rs11556218 T>G and rs4778889 T/C (IL16), miRNA-binding site polymorphisms in IL16, rs4464148 (SMAD7), rs6983267 (EGF), GSTT1, TACG haplotype (CTLA4), 1793G> A (MTHFR), Leu/Leu genotype of (EXO1), − 137 G/C (IL18), C/T genotype (XRCC3), I3434T (XRCC7), MGMT, C3435T (MDR1), ff genotype of FokI, 677CT+TT (MTHFR), G2677T/A (MDR1) and CRC. Increased risk has been observed in VDR ApaI genotype “aa”. Finally, the protective effect has been explored in the TACA haplotype (CTLA4). According to the findings, the genetic polymorphisms in the immunity-associated genes are related to the CRC amongst the Iranian patients. Therefore, more large-scale functional investigations are necessary for confirming the results.
Keywords: single nucleotide polymorphisms, colorectal cancer, Iran, biomarker
Introduction
Colorectal cancer (CRC) is a common third leading cause of death throughout the world, which can be prevented. It is the fifth most common cancer among men and the third most common cancer among women. The disease is more common in developed countries and accounts for 65% in these countries.1 CRC has been estimated to be the fourth most common cancer in Iran.2 The prevalence of CRC in different communities varies according to different environmental factors, human behaviors, and lifestyle. Aging, inflammatory bowel disease (IBD), family history, hereditary cancer syndromes, and lifestyle-related factors (eg, diabetes, inactive lifestyle, alcohol abuse, use of the red meat, and food with low fiber) are the most important factors increasing the risk of this malignancy.3 About 80% of CRC is caused by changes that are observed as chromosomal instability, aneuploidy, and early inactivation of adenomatous polyposis coli (APC) as found in the familial adenomatous polyposis (FAP). The remaining 15% is caused by disorders that lead to microsatellite instability and defects in the function of mismatch repair (MMR) genes such as hereditary non-polyposis colorectal cancer (HNPCC).4 Moreover, some studies found a relationship between genetic polymorphisms in individuals and an increase or decrease of their susceptibility to various cancers. Also, the genetic association studies on SNP have concentrated on the effect of single nucleotide polymorphisms on the candidate genes and cancer risk. The most important candidate genes include the genes involved in DNA repair and the immune system. SNPs can be found in coding and non-coding regions of the genes.5 Many SNPs do not affect the cell function; however, scientists believe that some of them can make a person susceptible to disease or affect their response to treatment. Therefore, it is difficult to find a relationship between diseases and SNPs with conventional methods because a gene may only play a minor role in the pathogenesis process. Individuals with specific SNPs or several SNPs may be more sensitive when exposed to carcinogens such as radiations. An SNP alone can increase the risk of cancer, but the presence of several polymorphic regions further increases the probability of cancer.6 It should be mentioned that this review has given a summary of the important genetic markers for the first time in Iranian patients with CRC (Table 1). Therefore, the mentioned genetic markers have been grouped in distinctive cellular procedures based on the respective cellular function (Figure 1).
 |  |  |
Table 1 All of the Studies Markers in CRC Susceptibility Among the Iranian Patients |
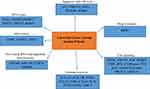 |
Figure 1 All the cellular processes which are studied in CRC progression among Iranian patients. |
SNPs in Genes Coding for Signaling
One of the proteins involved in the insulin signaling pathway is the insulin-like growth factor 1 (IGF1). This protein, on the one hand, stimulates cell division7 and on the other hand, prevents the programmed cell death (PCD).8 Those with elevated IGF1 levels are 2.5 times more likely to suffer from colorectal cancer.9 Numerous factors such as diet, lifestyle, genetic factors, and BMI are involved in controlling the IGF1 levels.10 However, due to the effects of IGF1 on the cell proliferation and differentiation of the large intestine cells in vitro and the biological system and its effect on the incidence of obesity and insulin resistance, researchers investigated its role in colorectal cancer.11 One of the proteins binding to the IGF1 is Insulin Growth Factor Binding Protein 3 (IGFBP3), which induces the programmed cell death through two pathways of p53- and IGF1-dependent. Moreover, some studies proved that the proteins binding to the IGF1, in particular, IGFBP3, play an important role in regulating the growth of the large intestine cells and nucleotide changes in the IGFBP3 gene are involved in gastrointestinal tumors.12 Other studies investigated the possible role of increased vitamin D in the reduction of the risk of CRC.13,14 Vitamin D activity depends on binding to the specific intracellular vitamin D receptor (VDR), whose nucleotide sequence in the human genome is known to be a highly polymorphic gene region. The function of this receptor represents a direct relationship with the amount of vitamin D active in the circulation and the values measured in vitro. VDR protein is known to facilitate tumorigenesis in the early stages of the disease because it increases in tumor tissues with the exception of its advanced stage.15 It is notable that Parathyroid hormone (PTH) works as one of the main regulators of the calcium homeostasis and modifies the expression of the proteins involved in the cell cycle in the CRC cells. In this regard, some research demonstrated higher serum levels of PTH in cases with CRC in comparison to the control group.16 Moreover, it has been indicated that PTH gene variants had a relationship with PTH and calcium serum levels.17 Calcium and calcium sensing receptor (CASR) have been considered to be the structured controllers of the colonocytes and calcium via signalling across CASR, which suppressed the proliferation of the normal colonocyte.18 Seemingly, CASR contributed to the suppression of tumor in CRC and had an essential contribution to maintain the calcium homeostasis.19
One of the studies in the field showed the fundamental involvement of epidermal growth factor (EGF) in tumor biology. EGF stimulated the cell proliferation, metastasis, invasion, angiogenesis as well as apoptosis inhibition.20 Also, several investigations confirmed that EGF and the respective receptor genes were over-expressed in different solid tumors, in particular, in carcinomas.21 Moreover, several replicative investigations proved that EGF 61A/G polymorphism could change risks for colon cancer.22 Another study demonstrated that EFNA1 was a glycosyl phosphatidyl inositol-linkage ligand with 205 amino acids binding to the receptor tyrosine kinase EphA2 at sites, in which the cell to cell contact occurred. The mentioned sites led to the contact-dependent bilateral signaling and played a significant role in the tumor neo-vascularization and development.23 Another study also showed that EFNA1 and its receptor EphA2 regulated the integrin-mediated adhesion, the cell proliferation, migration, and differentiation so that they have been regarded as the main mediators in developing and maintain various kinds of tumors.24 In fact, as stated in some studies, they contributed to several oncogenesis signaling pathways, including PI3K and MAP/ERK, and may affect the initiation and development of tumor.25,26 Finally, there has been a significant relationship between EFNA1 over-expression and TNM staging and the lymph node metastasis in the human gastric adenocarcinoma.27 According to the studies, SMAD7 has been considered to be a suppressive SMAD, and because of its contribution as one of the negative regulators of TGF-β signaling pathway, it enhances the anti-inflammatory impacts of TGF-β pathway.28 Hence, SMAD7 activities may notably decline TGF-β signaling and cause the enhanced risks of cancer.29 Concerning the genome-wide association studies (GWAS), researchers found a relationship of numerous loci and susceptibility to the CRC like diverse variants into SMAD7.30
Genetic Polymorphisms in the Immunity-Related Genes
As shown in one study, the IL1 gene has been considered to be situated on chromosome 2q14, which included 3 associated genes such as IL1A, IL1B, and IL1RN that encoded IL1a, IL1b, and IL1 receptor antagonist (IL1ra). IL1ra that is one of the anti-inflammatory cytokines would competitively bind to IL1 receptors and handles the inflammatory action of IL-1.31 On the other hand, the IL1RN gene possesses an 86-bp variable number of tandem repeats (VNTR) in the second intron. Several research indicated the enhanced risk of gastric cancer32 and CRC33 by IL1RN VNTR. Another study indicated that the transforming growth factor b (TGF-b) has been a cytokine, which has been used as the tumor inhibitor in the normal intestinal epithelium via suppression of the cell proliferation and induction of apoptosis.34 Also, multiple research suggested possible contribution of abnormality in the TGF-b pathway to oncogenesis especially to the colorectal carcinoma progression.35 Another study showed that TGFB1 level has been controlled genetically and researchers observed numerous polymorphisms in the TGFB1 gene, which affected the expression of TGF-b protein.36 According to some studies, CCL22 has been considered to be a Chemokine generated basically by macrophages. Moreover, Dendritic cells and several kinds of tumor cells could be used to secrete CCL22.37,38 Also, CCR4 which has been regarded as the prominent receptor of CCL22 is a member of the G-protein coupled receptor family of the proteins. Furthermore, researchers stated this receptor expression on the surface of multiple malignant cells.38,39 According to the last reports on the immune-suppressive contribution of CCR4 and CCL22 to numerous cancers, the CCL22 expression level has increased by gastrointestinal cells, in particular, under pathological conditions.40,41 Notably, the gene for cytotoxic T-lymphocyte antigen-4 (CTLA4) is a key gene that contributes to the immune response to different antigens. Investigations indicated the CTLA4 constitutive expression in the tumor cell lines at different intensities. Some of the also demonstrated the effects of CTLA4 gene polymorphism on the function and expression of CTLA4. Nonetheless, the most examined polymorphism of the CTLA4 gene has been reported to be an A to G substitution at position +49 in exon 1 but today researchers confirmed the effect of this SNP on the function and expression of the CTLA4 molecule.42
Another study showed that interleukin IL18 is a proinflammatory cytokine found in an inactive precursor in the normal gut mucosa, which could be quickly converted into a biologically active molecule via interleukine-1 beta converting enzyme.43 However, studies demonstrated a relationship between multiple proinflammatory gene products like IL18 and tumorgenesis, proposing the inflammation as one of the risk factors for developing cancer.44 Hence, IL18 as a proinflammatory cytokine contributed to the gastrointestinal inflammation possibly results in progressing cancer in the gastrointestinal tract. Moreover, IL18 production and/or activity could be changed by variations in the IL18 gene promoter and thus affects the genetic susceptibility to the development of various cancers.45 It is widely accepted that SDF-1 is a CXC chemokine, which binds to CXCR7 and CXCR4 receptors and contributes to the B and T lymphocytes homing and maturation,46 angiogenesis, regulating immunity, as well as the stem cell trafficking.47 In fact, it significantly contributes to the growth, development, and metastasis of various tumors like osteosarcoma48 and breast cancer.49 SDF-1 possesses a nucleotide transition from G to A (G→A) at position 801 in the 3′-untranslated region (SDF-1-G801A) in its β transcript that is also called SDF1-3′A.50 Thus, SDF-1-3′A could have a significant modulatory contribution via enhancing SDF-1 protein generation.51 Finally, this SNP of the SDF-1 gene had a relationship to the solid tumors. Another study indicated that the gene encoding IL16 cytokine contained eight exons spanning ~17 kb of genomic DNA situated at the chromosome 15q26.3 in the human genome.52 Multiple investigations emphasized the strong relationship of inflammatory parameters like pro-inflammatory cytokines with cancers.53 Moreover, the combined inflammatory cytokines generated by the epithelial cells of the colon and rectum in the tumor micro-environment could importantly contribute to cancer progression.54 PTGS2 is a pro-inflammatory and inducible enzyme, which converted arachidonic acid into prostaglandins.55 In fact, induction of the PTGS2 gene expression in the epithelial cells with increased growth has been observed so that there has been a relationship with the cancer invasion and development.56 However, there are growing documents of the relationship of PTGS2 polymorphisms with the risks of CRC.57,58 Therefore, a positive relationship between SNP and the increased risks of CRC has been established about the significance of PTGS2 enzyme in the inflammatory reaction that would be one of the crucial prerequisites for developing adenoma sot that this polymorphism position on the gene promoter region had a direct effect on the modulation of the gene expression and the rate of the enzyme generation.57–59
Genetic Polymorphisms in DNA Repair-Related Genes
One of the steps to repair DNA is to remove methyl from the O-6 atom of guanine created under alkylating agents. The presence of methylated guanine nucleotides at position 6 of the oxygen atom converts the G:C base pair to A: T, which is a transition mutation.60 The enzyme O6-methyl guanine methyltransferase (MGMT) is responsible for repairing this anomaly that removes the alkyl agent from the O6 atom position of the guanine nucleotide and maintains the structure of the original genome. Non-expression of this protein increases the risk of carcinogenicity and sensitivity to the methylating agents.61 Based on the studies, MGMT methylation occurs in the promoter region occurs in normal cells that soon become cancer cells.62 Also, MGMT gene silencing with an increase in the promoter region methylation of this gene has been observed in 20–40% of patients with CRC.62–65 Exonuclease 1 (EXO1) has been considered as one of the members of the RAD2 nuclease family with a significant contribution to the DNA replication, recombination, and mismatch repair.66 Also, the probable functional polymorphisms in EXO1 could involve in changing the CRC risks via effects on the repair activities of EXO1.67 Thus, this hypothesis would be reasonable that there is possibly a relationship between SNPs in EXO1 and CRC risks.
It has been also shown that RAD51 would play a significant role in the double-strand breaks (DSB) repair of DNA. In fact, the SNPs in this gene may affect the DNA repair potential and consequently the susceptibility to different tumors like CRC. In addition, RAD51 crucially contributes to the DSB repair across the homologous recombination (HR). It is notable that a major pathway to the DNA repair is the excision repair (ER) and double-strand break repair (DSBR). In fact, DSBR contained 2 mechanisms of homologous recombination (HR) and nonhomologous end joining (NHEJ). Therefore, different proteins and enzymes like XRCC3 and DNA-dependent protein kinase (DNA-PK) involve in each mechanism for repairing the injured DNA. Actually, the protein encoded by XRCC3 gene has been XRCC3 and DNA-PKcs has been the product of XRCC7 gene. Notably, the two proteins contributed to the DSBR mechanism; that is, DNA-PK in NHEJ and XRCC3 in HR.68 Recent researchers investigated the relationship between CRC and X-ray repair cross-complementing protein 3 (XRCC3) as the gene involved in the homologous recombination pathway.69–71 XRCC3 is an essential protein for chromosomal stability and cellular resistance to radiation and some chemical agents; however, in spite of its importance in repairing DNA. DSBs through homologous recombination pathway, there is not enough information of its biochemical properties and specific function.72 As a basic origin of dietary methyl groups, folate contributes importantly to the DNA methylation, repair, and synthesis. In addition, 5.10-methylene-tetrahydrofolate reductase (MTHFR) has been considered to be a major enzyme in the folate metabolism that performs irreversible conversion of 5.10-methylene-tetrahydrofolate into 5-methyl-tetrahydrofolate that would direct the folic acid pool towards re-methylation of homocysteine to methionine.73 It is notable that folate deficiency can result in the uracil mis incorporation and consequent DNA instability,74 retarded DNA repair capacities for oxidative or alkylating damage,75 and global and proto-oncogenic DNA hypomethylation.76 Thus, each effect contributes to carcinogenesis and the increased intake of folate has a relationship with lower risks of a number of cancers like CRC.77 Nonetheless, the relationship between folate and CRC has been incompatible with regard to the last studies. Moreover, there is a relationship between sufficient folate intake and the highly declined risk of CRC;78 however, there has been not a relationship with the endometrial cancer risks.79
SNPs in the Cell Cycle Genes
Some studies recognized approximately 14 SNPs in wild type TP53(p53) gene that can alter the p53 protein function.80,81 Pro72Arg (rs1042522) is a common SNP of the TP53 gene situated at the proline-rich domain of p53, which is crucial for normal p53 functions.82 Moreover, arginine (Arg) variant could induce apoptosis more rapidly and efficiently than proline (Pro) whereas Pro variant had a more acceptable function to induce the cycle arrest. The cancer risk could be enhanced by Pro72Arg SNP in the TP53 gene.83 One of the studies in the field showed the relationship of IL-16, CDKN2A (p16), RAF1, PTGER4, and ITGB4 with various cancers. CDKN2A has been considered to be a popular gene due to the respective impacts on pancreatic cancer.84 Moreover, ITGB4 has been categorized into the integrin protein group. The above proteins should provide the grounds for the cell-cell and cell-extracellular matrix adhesion. Because of such properties, the above proteins could contribute to cancer development.28 PTGER4 works as one of the negative feedback regulators of cell proliferation or rapid growth and thus the respective changes may cause the tumor progression.85 Therefore, variations in expressing the cancer-associated genes, CDKN2A (p16), RAF1, PTGER4, and ITGB4 amongst a population could apply negative or positive impacts on the individuals’ susceptibility to cancers.28,86-88
Non-Coding RNAs and Epigenetic Modification in CRC
According to a study in the field, the genetic polymorphism in the miRNA-binding region of the mRNA’s 3ʹ-UTR could lessen the miRNA- mRNA interactions, change expressing the target gene, and influence the individuals’ risks of disease.89 Numerous investigations demonstrated a potent contribution of rs1447295 polymorphism to susceptibility to cancers. Thus, this variant situated at the cancer susceptibility candidate 8 (CASC8) has been considered to be a long non-coding RNA (lncRNA) gene that would not code protein.90 CASC8 has been located adjacent to the Myc gene in the 8q24.1 region that is a popular gene desert consisting of several enhancer elements in the proximal of the MYC gene, associated CRC.91 Finally, the above enhancers modulate the MYC gene transcription via interacting with the CASC8 promoter.92
Polymorphisms of Anti-Oxidant Enzymes in CRC
Glutathione S-transferases (GSTs) that has been considered as one of the superfamilies’ of the dimeric Phase II metabolic enzymes contribute importantly to the cellular defense mechanism. Research showed a wide expression of sub-class GSTP1 in the normal human epithelial tissues and high overexpression in colon cancer.93 Moreover, GSTM1 and GSTT1 deficiency is related to higher risks of specific cancers.94,95 The epidemiologic investigations studying the relationship between variants in CYP and GST genes and colorectal neoplasia presented inconsistent outputs and thus there has been no agreement on their etiologic significance.96,97 However, a way to examine the protective contribution of GSTs would be to study the polymorphism in GSTM1, GSTT1, GSTP1, and CYP2E1 genes on the susceptibility to the CRC.98 Another study showed that P-glycoprotein (P-gp) that is a product of a multi-drug resistance gene (MDR1) would be one of the significant ATP-dependent membrane transporters that contributed to absorbing, distributing, and eliminating multiple medicines and works as the energy-dependent efflux pump exporting its substrates out of the cell.99 However, the most prominent contribution of P-gp would be the protection of the organism in opposition of xenobiotics and toxic compositions.100 Some studies demonstrated not less than 28 SNPs of MDR1 gene locus.100–102 For example, Hoffmeyer et al103 observed a silent polymorphism that has been related to P-gp expression. The observed polymorphism contained a C to T exchange at position 3435 in exon 26 of the MDR1 gene.
Microsatellite Instability in CRC
In 1993, it was found that the dysfunction of genes involved in the mismatch repair (MMR) pathway leads to microsatellite instability (MSI) in tumors with this defect. The cause of MSI is the presence of a mutation in the germline of the MMR system. The microsatellite sequence is scattered throughout the genome and consists of tandemly repeated sequences of 1–6 base pairs, which show very high polymorphism.104 In some tumors, half or more of the microsatellite become instable; a condition known as high-level microsatellite instability or MSI-High. Depending on the degree of instability, MSI is classified into three classes of MSI-High (MSI-H), MSI-Low (MSI-L), and MS-Stable (MSS). Microsatellite instability measurement is an excellent and fairly easy way to detect the deficiency of proteins of the MMR system.105 Among the molecular markers considered for the diagnosis and characterization of colorectal cancer, MSI has the advantage of providing promising information about the recurrence of this cancer. Colorectal cancer patients with MSI-High have multiple deletion/insertion mutations in at least two of the five DNA loci.106
Conclusions
Finding new prognostic factors, new biomarkers or pathologically modified characteristics is a never ending story and would be the subject of future research. Also, a personal approach to medicine (personalized medicine) has changed the field of oncology during the last decade. Tumor, Node, Metastasis (TNM) clinical and pathologic staging system is still the most important prognostic factor; however, in certain cancers and specific stages, it does not provide enough prognostic information and predictive information due to the heterogeneity of CRC. Therefore, new molecular objectives lead to the study of the biomarkers as the prognostic and predictive agents. Understanding the molecular mechanisms involved in the CRC development and metastatic stages will help us identify people at the highest risk of recurrence and find new tumor targets to prevent disease progression. Thus, one of the new biomarkers in CRC may be SNPs that affect the disease incidence and have the potential to be a prognostic and or predictive agent for daily clinical actions and decisions. However, it is important to identify the best and most relevant SNPs. According to Table 1, the presence of SNP in −800 G/A (TGFB1), Pro72Arg rs1042522 (TP53), rs4072111 (IL16), miRNA-binding site, CDKN2A (p16), RAF1, PTGER4, ITGB4, rs3135500 (NOD2), rs1368439 (IL12B), rs12953717 (SMAD7), −765G>C (PTGS2), rs4444903 (EGF), GSTM1, GSTP1, rs4359426 (CCL22), rs2228428 (CCR4), rs1447295 (CASC8), rs6983267 and rs10505477 (enetic variants), −607 C/A (IL18), rs6214 (IGF-I), rs3110697 (IGFBP-3), rs1052371 (INSR), (rs2289046) IRS2, methyl transferase 1, rs5277 (PTGS2/COX2), rs1801725 (CASR gene), rs6256 (PTH gene), Vitamin D receptor gene TaqI, codon 72 of the p53 gene (Arg72Pro), A1298C variants of MTHFR, miR-608 (rs4919510) and miR- 149 (rs2292832), SDF-1 gene at position 801 (G>A), C1236T (MDR1), rs12904 in the 3ʹ-UTR of ephrin A1, 135G>C (RAD51), A17893G (XRCC3) genes has no significant relationship with any types of the CRC, while there is a significant relationship between SNP in IL1RN* 2 (IL1ra), −509 C/T (TGFB1), rs11556218 T > G and rs4778889 T/C (IL16), miRNA-binding site polymorphisms in IL16, rs4464148 (SMAD7), rs6983267 (EGF), GSTT1, TACG haplotype (CTLA4), 1793G> A (MTHFR) Leu/Leu genotype of (EXO1), −137 G/C (IL18), C/T genotype (XRCC3), I3434T (XRCC7), O6-methylguanine-DNA methyltransferase, C3435T (MDR1), ff genotype of FokI, (MTHFR) 677CT+TT, G2677T/A (MDR1) and CRC. Increase risk has been observed in VDR ApaI genotype “aa”. Finally, the protective effect has been explored in TACA haplotype (CTLA4). According to the findings, the genetic polymorphisms in the immunity-associated genes related with the CRC amongst the Iranian Patients. Therefore, more large-scale functional investigations would be necessary for confirming the results.
Future Perspectives
Molecular pathological epidemiology (MPE) is an integrative discipline that combines epidemiology with molecular pathology. This field is structured around the core principle that diseases can be viewed as the product of profiles of exposomes, epigenomes, transcriptomes, proteomes, metabolomes, microbiomes, and interactomes and how they affect and are affected by macroenvironment and tissue microenvironment. This core principle, which is known as the unique disease principle, distinguish MPE from genome-wide association studies (GWAS).141 MPE helps researchers to gain valuable insights into the heterogeneity of diseases and produce epidemiologic data to further the study of molecular pathogenic mechanisms. With the wealth of biomedical data available to today’s researchers, they can use MPE to refine these data to gain a better understanding of the etiology and pathogenesis of diseases.142 MPE studies of CRC premalignant lesions can greatly contribute to personalized prevention, screening, and treatment of CRC cancer by offering valuable insights into the etiological factors of neoplastic initiation and progression and therefore the causes and risk factors of this cancer.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14(2):89–103. doi:10.5114/pg.2018.81072
2. Rafiemanesh H, Pakzad R, Abedi M, et al. Colorectal cancer in Iran: epidemiology and morphology trends. Excli j. 2016;15:738–744. doi:10.17179/excli2016-346
3. Gandomani HS, Yousefi SM, Aghajani M, et al. Colorectal cancer in the world: incidence, mortality and risk factors. Biomed Res Ther. 2017;4(10):1656–1675. doi:10.15419/bmrat.v4i10.372
4. Tariq K, Ghias K, Tariq K, Ghias K. Colorectal cancer carcinogenesis: a review of mechanisms. Cancer Biol Med. 2016;13(1):120–135. doi:10.20892/j.issn.2095-3941.2015.0103
5. Deng N, Zhou H, Fan H, Yuan Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 2017;8(66):110635. doi:10.18632/oncotarget.22372
6. Tan H. The association between gene SNPs and cancer predisposition: correlation or causality? EBioMedicine. 2017;16:8–9. doi:10.1016/j.ebiom.2017.01.047
7. Lahm H, Suardet L, Laurent P, et al. Growth regulation and co-stimulation of human colorectal cancer cell lines by insulin-like growth factor I, II and transforming growth factor α. Br J Cancer. 1992;65(3):341–346. doi:10.1038/bjc.1992.69
8. Remacle-Bonnet MM, Garrouste FL, Heller S, Andre F, Marvaldi JL, Pommier GJ. Insulin-like growth factor-I protects colon cancer cells from death factor-induced apoptosis by potentiating tumor necrosis factor alpha-induced mitogen-activated protein kinase and nuclear factor kappaB signaling pathways. Cancer Res. 2000;60(7):2007–2017.
9. Wei EK, Ma J, Pollak MN, et al. A prospective study of C-peptide, insulin-like growth factor-I, insulin-like growth factor binding protein-1, and the risk of colorectal cancer in women. Cancer Epidemiol Biomarkers Prev. 2005;14(4):850–855. doi:10.1158/1055-9965.EPI-04-0661
10. Ferry RJ
11. Morimoto LM, Newcomb PA, White E, Bigler J, Potter JD. Insulin-like growth factor polymorphisms and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14(5):1204–1211. doi:10.1158/1055-9965.EPI-04-0695
12. Allard JB, Duan C. IGF-binding proteins: why do they exist and why are there so many? Front Endocrinol (Lausanne). 2018;9:117. doi:10.3389/fendo.2018.00117
13. Jones G, Strugnell SA, DeLuca HF. Current understanding of the molecular actions of vitamin D. Physiol Rev. 1998;78(4):1193–1231. doi:10.1152/physrev.1998.78.4.1193
14. Sakulpipatsin W, Verasertniyom O, Nantiruj K, Totemchokchyakarn K, Lertsrisatit P, Janwityanujit S. Vitamin D receptor gene BsmI polymorphisms in Thai patients with systemic lupus erythematosus. Arthritis Res Ther. 2006;8(2):R48. doi:10.1186/ar1910
15. Peters U, McGlynn KA, Chatterjee N, et al. Vitamin D, calcium, and vitamin D receptor polymorphism in colorectal adenomas. Cancer Epidemiol Biomarkers Prev. 2001;10(12):1267–1274.
16. Fedirko V, Riboli E, Bueno-de-Mesquita HB, et al. Prediagnostic circulating parathyroid hormone concentration and colorectal cancer in the European prospective investigation into cancer and nutrition cohort. Cancer Epidemiol Biomarkers Prev. 2011;20(5):767–778. doi:10.1158/1055-9965.EPI-10-1212
17. Robinson-Cohen C, Lutsey PL, Kleber ME, et al. Genetic variants associated with circulating parathyroid hormone. J Am Soc Nephrol. 2017;28(5):1553–1565. doi:10.1681/ASN.2016010069
18. Whitfield JF. Calcium, calcium-sensing receptor and colon cancer. Cancer Lett. 2009;275(1):9–16. doi:10.1016/j.canlet.2008.07.001
19. Sarkar P, Kumar S. Calcium sensing receptor modulation for cancer therapy. Asian Pac J Cancer Prev. 2012;13(8):3561–3568. doi:10.7314/APJCP.2012.13.8.3561
20. Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19(3):183–232. doi:10.1016/1040-8428(94)00144-I
21. Siena S, Sartore-Bianchi A, Di Nicolantonio F, Balfour J, Bardelli A. Biomarkers predicting clinical outcome of epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer. J Natl Cancer Inst. 2009;101(19):1308–1324. doi:10.1093/jnci/djp280
22. Spindler KL, Nielsen JN, Ornskov D, Brandslund I, Jakobsen A. Epidermal growth factor (EGF) A61G polymorphism and EGF gene expression in normal colon tissue from patients with colorectal cancer. Acta Oncol. 2007;46(8):1113–1117. doi:10.1080/02841860701338853
23. Nakamura R, Kataoka H, Sato N, et al. EPHA2/EFNA1 expression in human gastric cancer. Cancer Sci. 2005;96(1):42–47. doi:10.1111/j.1349-7006.2005.00007.x
24. Beauchamp A, Debinski W. Ephs and ephrins in cancer: ephrin-A1 signalling. Semin Cell Dev Biol. 2012;23(1):109–115. doi:10.1016/j.semcdb.2011.10.019
25. Pandey A, Lazar DF, Saltiel AR, Dixit VM. Activation of the Eck receptor protein tyrosine kinase stimulates phosphatidylinositol 3-kinase activity. J Biol Chem. 1994;269(48):30154–30157.
26. Pratt RL, Kinch MS. Activation of the EphA2 tyrosine kinase stimulates the MAP/ERK kinase signaling cascade. Oncogene. 2002;21(50):7690–7699. doi:10.1038/sj.onc.1205758
27. Yuan WJ, Ge J, Chen ZK, et al. Over-expression of EphA2 and EphrinA-1 in human gastric adenocarcinoma and its prognostic value for postoperative patients. Dig Dis Sci. 2009;54(11):2410–2417. doi:10.1007/s10620-008-0649-4
28. Brendle A, Lei H, Brandt A, et al. Polymorphisms in predicted microRNA-binding sites in integrin genes and breast cancer: ITGB4 as prognostic marker. Carcinogenesis. 2008;29(7):1394–1399. doi:10.1093/carcin/bgn126
29. Akbari Z, Safari-Alighiarloo N, Haghighi MM, et al. Lack of influence of the SMAD7 gene rs2337107 polymorphism on risk of colorectal cancer in an Iranian population. Asian Pac J Cancer Prev. 2014;15(11):4437–4441. doi:10.7314/APJCP.2014.15.11.4437
30. Mias GI, Snyder M. Personal genomes, quantitative dynamic omics and personalized medicine. Quant Biol. 2013;1(1):71–90. doi:10.1007/s40484-013-0005-3
31. Dinarello CA. The interleukin-1 family: 10 years of discovery. FASEB J. 1994;8(15):1314–1325. doi:10.1096/fasebj.8.15.8001745
32. El-Omar EM, Carrington M, Chow WH, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404(6776):398–402. doi:10.1038/35006081
33. Lurje G, Hendifar AE, Schultheis AM, et al. Polymorphisms in interleukin 1 beta and interleukin 1 receptor antagonist associated with tumor recurrence in stage II colon cancer. Pharmacogenet Genomics. 2009;19(2):95–102. doi:10.1097/FPC.0b013e32831a9ad1
34. Pasche B. Role of transforming growth factor beta in cancer. J Cell Physiol. 2001;186(2):153–168. doi:10.1002/1097-4652(200002)186:2<153::AID-JCP1016>3.0.CO;2-J
35. Xu Y, Pasche B. TGF-β signaling alterations and susceptibility to colorectal cancer. Hum Mol Genet. 2007;16(R1):R14–R20. doi:10.1093/hmg/ddl486
36. Chung SJ, Kim JS, Jung HC, Song IS. Transforming growth factor‐β1‐509T reduces risk of colorectal cancer, but not adenoma in Koreans. Cancer Sci. 2007;98(3):401–404. doi:10.1111/j.1349-7006.2007.00401.x
37. Shimauchi T, Imai S, Hino R, Tokura Y. Production of thymus and activation-regulated chemokine and macrophage-derived chemokine by CCR4+ adult T-cell leukemia cells. Clin Cancer Res. 2005;11(6):2427–2435. doi:10.1158/1078-0432.CCR-04-0491
38. Mizukami Y, Kono K, Kawaguchi Y, et al. CCL17 and CCL22 chemokines within tumor microenvironment are related to accumulation of Foxp3+ regulatory T cells in gastric cancer. Int j Cancer. 2008;122(10):2286–2293. doi:10.1002/ijc.23392
39. Ishida T, Inagaki H, Utsunomiya A, et al. CXC chemokine receptor 3 and CC chemokine receptor 4 expression in T-cell and NK-cell lymphomas with special reference to clinicopathological significance for peripheral T-cell lymphoma, unspecified. Clin Cancer Res. 2004;10(16):5494–5500. doi:10.1158/1078-0432.CCR-04-0371
40. Yang S, Eckmann L, Panja A, Kagnoff MF. Differential and regulated expression of CXC, CC, and C-chemokines by human colon epithelial cells. Gastroenterology. 1997;113(4):1214–1223. doi:10.1053/gast.1997.v113.pm9322516
41. Eckmann L, Jung HC, Schurer-Maly C, Panja A, Morzycka-Wroblewska E, Kagnoff MF. Differential cytokine expression by human intestinal epithelial cell lines: regulated expression of interleukin 8. Gastroenterology. 1993;105(6):1689–1697. doi:10.1016/0016-5085(93)91064-O
42. Vaidya B, Imrie H, Perros P, et al. The cytotoxic T lymphocyte antigen-4 is a major Graves’ disease locus. Hum Mol Genet. 1999;8(7):1195–1199. doi:10.1093/hmg/8.7.1195
43. Pages F, Berger A, Lebel-Binay S, et al. Proinflammatory and antitumor properties of interleukin-18 in the gastrointestinal tract. Immunol Lett. 2000;75(1):9–14. doi:10.1016/S0165-2478(00)00285-6
44. Aggarwal BB, Shishodia S, Sandur SK, Pandey MK, Sethi G. Inflammation and cancer: how hot is the link? Biochem Pharmacol. 2006;72(11):1605–1621. doi:10.1016/j.bcp.2006.06.029
45. Giedraitis V, He B, Huang WX, Hillert J. Cloning and mutation analysis of the human IL-18 promoter: a possible role of polymorphisms in expression regulation. J Neuroimmunol. 2001;112(1–2):146–152. doi:10.1016/S0165-5728(00)00407-0
46. Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J Exp Med. 1996;184(3):1101–1109. doi:10.1084/jem.184.3.1101
47. Sun X, Cheng G, Hao M, et al. CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010;29(4):709–722. doi:10.1007/s10555-010-9256-x
48. Perissinotto E, Cavalloni G, Leone F, et al. Involvement of chemokine receptor 4/stromal cell-derived factor 1 system during osteosarcoma tumor progression. Clin Cancer Res. 2005;11(2 Pt 1):490–497.
49. Luker KE, Luker GD. Functions of CXCL12 and CXCR4 in breast cancer. Cancer Lett. 2006;238(1):30–41. doi:10.1016/j.canlet.2005.06.021
50. Watanabe MA, de Oliveira Cavassin GG, Orellana MD, et al. SDF-1 gene polymorphisms and syncytia induction in Brazilian HIV-1 infected individuals. Microb Pathog. 2003;35(1):31–34. doi:10.1016/S0882-4010(03)00088-3
51. Winkler C, Modi W, Smith MW, et al. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC). Science. 1998;279(5349):389–393. doi:10.1126/science.279.5349.389
52. Kim HS. Assignment of human interleukin 16 (IL16) to chromosome 15q26.3 by radiation hybrid mapping. Cytogenet Cell Genet. 1999;84(1–2):93. doi:10.1159/000015224
53. Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi:10.1038/nature01322
54. Sheu BC, Chang WC, Cheng CY, Lin HH, Chang DY, Huang SC. Cytokine regulation networks in the cancer microenvironment. Front Biosci. 2008;13(13):6255–6268. doi:10.2741/3152
55. Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, DuBois RN. Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology. 1994;107(4):1183–1188. doi:10.1016/0016-5085(94)90246-1
56. Trifan OC, Hla T. Cyclooxygenase-2 modulates cellular growth and promotes tumorigenesis. J Cell Mol Med. 2003;7(3):207–222. doi:10.1111/j.1582-4934.2003.tb00222.x
57. Cox D, Pontes C, Guinó E, et al. Polymorphisms in prostaglandin synthase 2/cyclooxygenase 2 (PTGS2/COX2) and risk of colorectal cancer. Br J Cancer. 2004;91(2):339–343. doi:10.1038/sj.bjc.6601906
58. Goodman JE, Bowman ED, Chanock SJ, Alberg AJ, Harris CC. Arachidonate lipoxygenase (ALOX) and cyclooxygenase (COX) polymorphisms and colon cancer risk. Carcinogenesis. 2004;25(12):2467–2472. doi:10.1093/carcin/bgh260
59. Zhu W, Wei B, Shan X, Liu P. 765G> C and 8473T> C polymorphisms of COX-2 and cancer risk: a meta-analysis based on 33 case–control studies. Mol Biol Rep. 2010;37(1):277–288. doi:10.1007/s11033-009-9685-1
60. Aquilina G, Biondo R, Dogliotti E, Meuth M, Bignami M. Expression of the endogenous O6-methylguanine-DNA-methyltransferase protects Chinese hamster ovary cells from spontaneous G:C to A:T transitions. Cancer Res. 1992;52(23):6471–6475.
61. Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4(4):296–307. doi:10.1038/nrc1319
62. Nagasaka T, Sharp GB, Notohara K, et al. Hypermethylation of O6-methylguanine-DNA methyltransferase promoter may predict nonrecurrence after chemotherapy in colorectal cancer cases. Clin Cancer Res. 2003;9(14):5306–5312.
63. Esteller M, Toyota M, Sanchez-Cespedes M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60(9):2368–2371.
64. Esteller M, Risques RA, Toyota M, et al. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61(12):4689–4692.
65. Whitehall VL, Walsh MD, Young J, Leggett BA, Jass JR. Methylation of O-6-methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low-level DNA microsatellite instability. Cancer Res. 2001;61(3):827–830.
66. Schmutte C, Marinescu RC, Sadoff MM, Guerrette S, Overhauser J, Fishel R. Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res. 1998;58(20):4537–4542.
67. Tsai M-H, Tseng H-C, Liu C-S, et al. Interaction of Exo1 genotypes and smoking habit in oral cancer in Taiwan. Oral Oncol. 2009;45(9):e90–e94. doi:10.1016/j.oraloncology.2009.03.011
68. Mohiuddin IS, Kang MH. DNA-PK as an Emerging Therapeutic Target in Cancer. Front Oncol. 2019;9:635. doi:10.3389/fonc.2019.00635
69. Namazi A, Abedinzadeh M, Nourbaksh P, Neamatzadeh H. Association between the XRCC3 Thr241Met polymorphism and risk of colorectal cancer: a meta analysis of 5193 cases and 6645 controls. Asian Pac J Cancer Prev. 2015;16(6):2263–2268. doi:10.7314/APJCP.2015.16.6.2263
70. Krupa R, Sliwinski T, Wisniewska-Jarosinska M, et al. Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer–a case control study. Mol Biol Rep. 2011;38(4):2849–2854. doi:10.1007/s11033-010-0430-6
71. Zhao Y, Deng X, Wang Z, Wang Q, Liu Y. Genetic polymorphisms of DNA repair genes XRCC1 and XRCC3 and risk of colorectal cancer in Chinese population. Asian Pac J Cancer Prev. 2012;13(2):665–669. doi:10.7314/APJCP.2012.13.2.665
72. Alayev A, Salamon RS, Manna S, Schwartz NS, Berman AY, Holz MK. Estrogen induces RAD51C expression and localization to sites of DNA damage. Cell Cycle. 2016;15(23):3230–3239. doi:10.1080/15384101.2016.1241927
73. Kim Y-I. Folate and carcinogenesis: evidence, mechanisms, and implications. J Nutr Biochem. 1999;10(2):66–88. doi:10.1016/S0955-2863(98)00074-6
74. Wainfan E, Poirier LA. Methyl groups in carcinogenesis: effects on DNA methylation and gene expression. Cancer Res. 1992;52(7Supplement):2071s–2077s.
75. Sanjoaquin MA, Allen N, Couto E, Roddam AW, Key TJ. Folate intake and colorectal cancer risk: a meta‐analytical approach. Int J Cancer. 2005;113(5):825–828. doi:10.1002/ijc.20648
76. Shrubsole MJ, Jin F, Dai Q, et al. Dietary folate intake and breast cancer risk: results from the shanghai breast cancer study. Cancer Res. 2001;61(19):7136–7141.
77. McCann SE, Freudenheim JL, Marshall JR, Brasure JR, Swanson MK, Graham S. Diet in the epidemiology of endometrial cancer in western New York (United States). Cancer Causes Control. 2000;11(10):965–974. doi:10.1023/A:1026551309873
78. Negri E, La Vecchia C, Franceschi S, Levi F, Parazzini F. Intake of selected micronutrients and the risk of endometrial carcinoma. Cancer. 1996;77(5):917–923. doi:10.1002/(SICI)1097-0142(19960301)77:5<917::AID-CNCR17>3.0.CO;2-6
79. Jain MG, Howe GR, Rohan TE. Nutritional factors and endometrial cancer in Ontario, Canada. Cancer Control. 2000;7(3):288–296. doi:10.1177/107327480000700312
80. Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1(3):233–240. doi:10.1038/35106009
81. Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19(6):607–614. doi:10.1002/humu.10081
82. Dumont P, Leu JI, Della Pietra AC
83. Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19(2):1092–1100. doi:10.1128/MCB.19.2.1092
84. McWilliams RR, Wieben ED, Rabe KG, et al. Prevalence of CDKN2A mutations in pancreatic cancer patients: implications for genetic counseling. Eur J Hum Genet. 2011;19(4):472–478. doi:10.1038/ejhg.2010.198
85. Murn J, Alibert O, Wu N, Tendil S, Gidrol X. Prostaglandin E2 regulates B cell proliferation through a candidate tumor suppressor, Ptger4. J Exp Med. 2008;205(13):3091–3103. doi:10.1084/jem.20081163
86. Benjamin D, Moroni C. mRNA stability and cancer: an emerging link? Expert Opin Biol Ther. 2007;7(10):1515–1529. doi:10.1517/14712598.7.10.1515
87. Xiong F, Wu C, Chang J, et al. Genetic variation in an miRNA-1827 binding site in MYCL1 alters susceptibility to small-cell lung cancer. Cancer Res. 2011;71(15):5175–5181. doi:10.1158/0008-5472.CAN-10-4407
88. Gao LB, Rao L, Wang YY, et al. The association of interleukin-16 polymorphisms with IL-16 serum levels and risk of colorectal and gastric cancer. Carcinogenesis. 2009;30(2):295–299. doi:10.1093/carcin/bgn281
89. Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29(7):1306–1311. doi:10.1093/carcin/bgn116
90. Gudmundsson J, Sulem P, Manolescu A, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39(5):631–637. doi:10.1038/ng1999
91. Sotelo J, Esposito D, Duhagon MA, et al. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci U S A. 2010;107(7):3001–3005. doi:10.1073/pnas.0906067107
92. Kim T, Cui R, Jeon YJ, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc Natl Acad Sci U S A. 2014;111(11):4173–4178. doi:10.1073/pnas.1400350111
93. Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286(5439):487–491. doi:10.1126/science.286.5439.487
94. Rebbeck TR. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol Biomarkers Prev. 1997;6(9):733–743.
95. Strange RC, Lear JT, Fryer AA. Glutathione S-transferase polymorphisms: influence on susceptibility to cancer. Chem Biol Interact. 1998;111–112:351–364. doi:10.1016/S0009-2797(97)00172-5
96. Bailey LR, Roodi N, Verrier CS, Yee CJ, Dupont WD, Parl FF. Breast cancer and CYPIA1, GSTM1, and GSTT1 polymorphisms: evidence of a lack of association in Caucasians and African Americans. Cancer Res. 1998;58(1):65–70.
97. Saadat I, Saadat M. Glutathione S-transferase M1 and T1 null genotypes and the risk of gastric and colorectal cancers. Cancer Lett. 2001;169(1):21–26. doi:10.1016/S0304-3835(01)00550-X
98. Pemble S, Schroeder KR, Spencer SR, et al. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J. 1994;300(Pt 1):271–276. doi:10.1042/bj3000271
99. Arceci RJ. Clinical significance of P-glycoprotein in multidrug resistance malignancies. Blood. 1993;81(9):2215–2222. doi:10.1182/blood.V81.9.2215.2215
100. Jamroziak K, Mlynarski W, Balcerczak E, et al. Functional C3435T polymorphism of MDR1 gene: an impact on genetic susceptibility and clinical outcome of childhood acute lymphoblastic leukemia. Eur J Haematol. 2004;72(5):314–321. doi:10.1111/j.1600-0609.2004.00228.x
101. Ito S, Ieiri I, Tanabe M, Suzuki A, Higuchi S, Otsubo K. Polymorphism of the ABC transporter genes, MDR1, MRP1 and MRP2/cMOAT, in healthy Japanese subjects. Pharmacogenetics. 2001;11(2):175–184. doi:10.1097/00008571-200103000-00008
102. Kim RB, Leake BF, Choo EF, et al. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther. 2001;70(2):189–199. doi:10.1067/mcp.2001.117412
103. Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97(7):3473–3478. doi:10.1073/pnas.97.7.3473
104. Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(6):2073–2087.e2073. doi:10.1053/j.gastro.2009.12.064
105. Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol. 2015;16(7):30. doi:10.1007/s11864-015-0348-2
106. Nojadeh JN, Behrouz Sharif S, Sakhinia E. Microsatellite instability in colorectal cancer. EXCLI j. 2018;17:159–168. doi:10.17179/excli2017-948
107. Abbasian MH, Abbasi B, Ansarinejad N, et al. Association of interleukin-1 gene polymorphism with risk of gastric and colorectal cancers in an Iranian population. Iran J Immunol. 2018;15(4):321–328. doi:10.22034/IJI.2018.39401
108. Amirghofran Z, Jalali SA, Ghaderi A, Hosseini SV. Genetic polymorphism in the transforming growth factor beta1 gene (−509 C/T and −800 G/A) and colorectal cancer. Cancer Genet Cytogenet. 2009;190(1):21–25. doi:10.1016/j.cancergencyto.2008.11.010
109. Asadi M, Shanehbandi D, Zarintan A, et al. TP53 gene pro72Arg (rs1042522) single nucleotide polymorphism as not a risk factor for colorectal cancer in the iranian azari population. Asian Pac J Cancer Prev. 2017;18(12):3423–3427. doi:10.22034/APJCP.2017.18.12.3423
110. Azimzadeh P, Romani S, Mohebbi SR, et al. Interleukin-16 (IL-16) gene polymorphisms in Iranian patients with colorectal cancer. J Gastrointestin Liver Dis. 2011;20(4):371–376.
111. Azimzadeh P, Romani S, Mohebbi SR, et al. Association of polymorphisms in microRNA-binding sites and colorectal cancer in an Iranian population. Cancer Genet. 2012;205(10):501–507. doi:10.1016/j.cancergen.2012.05.013
112. Chaleshi V, Tajali R, Savabkar S, et al. Lack of Association between NOD2 rs3135500 and IL12B rs1368439 microRNA Binding Site SNPs and Colorectal Cancer Susceptibility in an Iranian Population. MicroRNA. 2016;5(2):152–156. doi:10.2174/2211536605666160715151535
113. Damavand B, Derakhshani S, Saeedi N, et al. Intronic polymorphisms of the SMAD7 gene in association with colorectal cancer. Asian Pac J Cancer Prev. 2015;16(1):41–44. doi:10.7314/APJCP.2015.16.1.41
114. Daraei A, Salehi R, Mohamadhashem F. PTGS2 (COX2) −765G>C gene polymorphism and risk of sporadic colorectal cancer in Iranian population. Mol Biol Rep. 2012;39(5):5219–5224. doi:10.1007/s11033-011-1319-8
115. Daraei A, Salehi R, Salehi M, et al. Effect of rs6983267 polymorphism in the 8q24 region and rs4444903 polymorphism in EGF gene on the risk of sporadic colorectal cancer in Iranian population. Med Oncol. 2012;29(2):1044–1049. doi:10.1007/s12032-011-9980-2
116. Dastjerdi MN. TP53 codon 72 polymorphism and P53 protein expression in colorectal cancer specimens in Isfahan. Acta Med Iran. 2011;49(2):71–77.
117. Ebrahimkhani S, Asgharian AM, Nourinaier B, et al. Association of GSTM1, GSTT1, GSTP1 and CYP2E1 single nucleotide polymorphisms with colorectal cancer in Iran. Pathol Oncol Res. 2012;18(3):651–656. doi:10.1007/s12253-011-9490-8
118. Erfani N, Ahrari S, Ahrari I, Hosseini SV. CCR4 C1014T and CCL22 C16A genetic variations in the Iranian patients with colorectal adenocarcinoma. Iran J Allergy Asthma Immunol. 2014;13(6):440–446.
119. Hadinia A, Hossieni SV, Erfani N, Saberi-Firozi M, Fattahi MJ, Ghaderi A. CTLA-4 gene promoter and exon 1 polymorphisms in Iranian patients with gastric and colorectal cancers. J Gastroenterol Hepatol. 2007;22(12):2283–2287. doi:10.1111/j.1440-1746.2007.04862.x
120. Haerian MS, Haerian BS, Molanaei S, et al. Lack of association of CASC8 rs1447295 with colorectal cancer in Iranian population: a multicenter case-control study. Gene. 2017;634:74–76. doi:10.1016/j.gene.2017.08.042
121. Haerian MS, Haerian BS, Rooki H, et al. Association of 8q24.21 rs10505477-rs6983267 haplotype and age at diagnosis of colorectal cancer. Asian Pac J Cancer Prev. 2014;15(1):369–374. doi:10.7314/APJCP.2014.15.1.369
122. Haghighi MM, Mohebbi SR, Najjar Sadeghi R, Vahedi M, Ghiasi S, Zali MR. Association between the 1793G> A MTHFR polymorphism and sporadic colorectal cancer in Iran. Asian Pac J Cancer Prev. 2008;9(4):659–662.
123. Haghighi MM, Taleghani MY, Mohebbi SR, et al. Impact of EXO1 polymorphism in susceptibility to colorectal cancer. Genet Test Mol Biomarkers. 2010;14(5):649–652. doi:10.1089/gtmb.2010.0034
124. Haghshenas MR, Hosseini SV, Mahmoudi M, Saberi-Firozi M, Farjadian S, Ghaderi A. IL-18 serum level and IL-18 promoter gene polymorphism in Iranian patients with gastrointestinal cancers. J Gastroenterol Hepatol. 2009;24(6):1119–1122. doi:10.1111/j.1440-1746.2009.05791.x
125. Karimi K, Mahmoudi T, Karimi N, et al. Is there an association between variants in candidate insulin pathway genes IGF-I, IGFBP-3, INSR, and IRS2 and risk of colorectal cancer in the Iranian population? Asian Pac J Cancer Prev. 2013;14(9):5011–5016. doi:10.7314/APJCP.2013.14.9.5011
126. Khatami F, Noorinayer B, Mohebi SR, et al. Effects of amino acid substitution polymorphisms of two DNA methyltransferases on susceptibility to sporadic colorectal cancer. Asian Pac J Cancer Prev. 2009;10(6):1183–1188.
127. Khedri A, Nejat-Shokouhi A, Salek R, et al. Association of the colorectal cancer and MDR1 gene polymorphism in an Iranian population. Mol Biol Rep. 2011;38(5):2939–2943. doi:10.1007/s11033-010-9957-9
128. Khorshidi F, Haghighi MM, Nazemalhosseini Mojarad E, et al. The prostaglandin synthase 2/cyclooxygenase 2 (PTGS2/COX2) rs5277 polymorphism does not influence risk of colorectal cancer in an Iranian population. Asian Pac J Cancer Prev. 2014;15(8):3507–3511. doi:10.7314/APJCP.2014.15.8.3507
129. Mahmoudi T, Karimi K, Arkani M, et al. Parathyroid hormone gene rs6256 and calcium sensing receptor gene rs1801725 variants are not associated with susceptibility to colorectal cancer in Iran. Asian Pac J Cancer Prev. 2014;15(15):6035–6039. doi:10.7314/APJCP.2014.15.15.6035
130. Mahmoudi T, Mohebbi SR, Pourhoseingholi MA, Fatemi SR, Zali MR. Vitamin D receptor gene ApaI polymorphism is associated with susceptibility to colorectal cancer. Dig Dis Sci. 2010;55(7):2008–2013. doi:10.1007/s10620-009-0989-8
131. Mehrzad J, Dayyani M, Khorasani ME. Polymorphisms of XRCC3 and XRCC7 and colorectal cancer risk in khorasan Razavi Province, Iran. Asian Pac J Cancer Prev. 2019;20(7):2153–2158. doi:10.31557/APJCP.2019.20.7.2153
132. Mojtahedi Z, Haghshenas MR, Hosseini SV, Fattahi MJ, Ghaderi A. p 53 codon 72 polymorphism in stomach and colorectal adenocarcinomas in Iranian patients. Indian J Cancer. 2010;47(1):31–34. doi:10.4103/0019-509X.58856
133. Moossavi M, Parsamanesh N, Mohammadoo-Khorasani M, et al. Positive correlation between vitamin D receptor gene FokI polymorphism and colorectal cancer susceptibility in South-Khorasan of Iran. J Cell Biochem. 2018;119(10):8190–8194. doi:10.1002/jcb.26826
134. Naghibalhossaini F, Mokarram P, Khalili I, et al. MTHFR C677T and A1298C variant genotypes and the risk of microsatellite instability among Iranian colorectal cancer patients. Cancer Genet Cytogenet. 2010;197(2):142–151. doi:10.1016/j.cancergencyto.2009.11.014
135. Parsafar S, Hematti S, Ghorbani F, Safari F, Tavassoli M. Polymorphic GT dinucleotide repeat in the PIK3CA gene and risk of colorectal cancer. Cancer Biomark. 2015;15(4):397–403. doi:10.3233/CBM-150487
136. Ranjbar R, Chaleshi V, Aghdaei HA, Morovvati S. Investigating the Association Between miR-608 rs4919510 and miR-149 rs2292832 with Colorectal Cancer in Iranian Population. MicroRNA. 2018;7(2):100–106. doi:10.2174/2211536607666180206145540
137. Razmkhah M, Ghaderi A. SDF-1alpha G801A polymorphism in Southern Iranian patients with colorectal and gastric cancers. Indian J Gastroenterol. 2013;32(1):28–31. doi:10.1007/s12664-012-0283-0
138. Samanian S, Mahjoubi F, Mahjoubi B, Mirzaee R, Azizi R. MDR1 gene polymorphisms: possible association with its expression and clinicopathology characteristics in colorectal cancer patients. Asian Pac J Cancer Prev. 2011;12(11):3141–3145.
139. Simonian M, Mosallaei M, Khosravi S, Salehi R. rs12904 polymorphism in the 3ʹ-untranslated region of ephrin A1 ligand and the risk of sporadic colorectal cancer in the Iranian population. J Cancer Res Ther. 2019;15(1):15–19. doi:10.4103/jcrt.JCRT_766_17
140. Yazdanpanahi N, Salehi R, Kamali S. RAD51 135G>C polymorphism and risk of sporadic colorectal cancer in Iranian population. J Cancer Res Ther. 2018;14(3):614–618. doi:10.4103/0973-1482.183558
141. Ogino S, Lochhead P, Chan AT, et al. Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease. Mod Pathol. 2013;26(4):465–484.
142. Hamada T, Keum N, Nishihara R, Ogino S. Molecular pathological epidemiology`ew developing frontiers of big data science to study etiologies and pathogenesis. J Gastroenterol. 2017;52(3):265–275. doi:10.1007/s00535-016-1272-3
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
