Back to Journals » Infection and Drug Resistance » Volume 11
Single-nucleotide polymorphisms related to fluoroquinolone and aminoglycoside resistance in Mycobacterium avium isolates
Received 27 December 2017
Accepted for publication 12 February 2018
Published 9 April 2018 Volume 2018:11 Pages 515—521
DOI https://doi.org/10.2147/IDR.S160899
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sahil Khanna
Video abstract presented by Pang et al.
Views: 273
Hui Pang,1,2 Kanglin Wan,3 Lin Wei1
1Department of Immunology, Hebei Medical University, Shijiazhuang, Hebei, China; 2Department of Immunology, Changzhi Medical College, Changzhi, Shanxi, China; 3State Key Laboratory for Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
Objective: The relationships between fluoroquinolone and aminoglycoside resistance and single-nucleotide polymorphisms (SNPs) in gyrA, gyrB, and rpsL genes were investigated in 95 clinical isolates of Mycobacterium avium from China.
Methods: Fluoroquinolone and aminoglycoside resistance were determined by the broth microdilution method. GyrA, gyrB, and rpsL were sequenced, SNPs were identified, and the corresponding amino acid mutations were recorded.
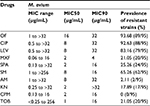
Results: The M. avium isolates displayed high levels of ofloxacin (93.68%), ciprofloxacin (92.63%), and streptomycin (65.26%) resistance. Moxifloxacin (18.95%) and amikacin (2.11%) were highly active against the strains. Fluoroquinolone resistance involving gyrA and gyrB gene mutations was identified. For gyrA, the most frequent SNPs were T→C (71/95, 74.74%), followed by A→G (64/95, 67.37%) and T→C (62/95, 65.26%). The amino acid mutations occurred mainly at Gly2444Asp (GGT→GAT) (20/95, 21.05%), Ala2445Ser (GCC→TCC) (20/95, 21.05%), Ala2447Val (GCC→GTC) (20/95, 21.05%), Val2449Ile (GTC→ATC) (20/95, 21.05%), and Glu2450Gln (GAA→CAA) (20/95, 21.05%). Prominent SNPs in gyrB included A→C (69/95, 72.63%), C→T (51/95, 53.68%), and T→G (29/95, 30.53%), and their amino acid substitutions were Ile2160Val (ATT→GTT) (21/95, 22.11%), Ile2160Met (ATT→ATG) (20/95, 21.05%), and Ile2273Leu (ATC→CTC) (11/95, 11.58%). Among the strains with aminoglycoside resistance, SNPs in rpsL were identified mostly at position G→A (73/95, 76.84%). G→C (21/95, 22.11%) was commonly seen. The amino acid mutations primarily involved Ala1539985Thr (GCC→ACC) (19/95, 20.00%), His1539992Asp (CAC→GAC) (19/95, 20.00%), and Gln1539983Glu (CAG→GAG) (18/95, 18.95%).
Conclusion: Our study provides valuable information that could be used for the future diagnosis and treatment of M. avium disease.
Keywords: Mycobacterium avium, drug resistance, single-nucleotide polymorphism, amino acid mutation, minimum inhibitory concentration
Introduction
Mycobacterium avium was first isolated from chickens in 1933 as a cavitary disease, which was similar to tuberculosis.1 This species is the most common cause of nontuberculosis mycobacterial infections in humans, especially respiratory system diseases.1 The incidence and prevalence of M. avium lung diseases have been increasing worldwide.2 M. avium lung disease often affects elderly people with chronic lung diseases and may be the manifestation of a complex genetic disorder determined by the interactions of multiple genes as well as environmental factors.2,3 It is difficult to treat and requires carefully individualized analysis of the risks and benefits of treatment.2,3 Despite extensive research into drug resistance in Mycobacterium tuberculosis, few studies have investigated the molecular mechanisms underlying drug susceptibility in M. avium.4–6 There have been continuous efforts to address problems, such as drug resistance to conventional antituberculosis agents, and research using M. avium could be beneficial.
DNA gyrase, the primary target of fluoroquinolone drugs, relaxes DNA supercoiling ahead of the DNA helicase–DNA replication complex. DNA gyrase is encoded by the gyrA and gyrB genes.7,8 In contrast, aminoglycosides inhibit protein synthesis by binding to the ribosome near the A site. The primary mechanism of acquires resistance to aminoglycosides in mycobacteria is based on modification of the 30S subunit of the ribosome as the drug target.9,10 This modification is caused by mutations, often in the rpsL gene, which encodes the S12 ribosomal protein.11 Other mutations associated with drug resistance involve single-nucleotide polymorphism (SNP) changes to a single base at a specific position in the genome. The correlation between SNPs and phenotypic diversity has been established for some mycobacterial species, encouraging further correlation analyses to be done to distinguish the SNPs among the strains.11,12
In this study, the broth microdilution method recommended by the Clinical and Laboratory Standards Institute was used to test the drug sensitivity including 10 antimicrobial agents of 95 M. avium clinical isolates collected from five Chinese provinces between 2005 and 2012.12 The SNP analysis was performed to determine the mutation characteristics of the strains carrying fluoroquinolone and aminoglycoside resistance. The results of this study should be helpful for improving the diagnosis and treatment of M. avium infections in the clinical work.
Materials and methods
Strains
All 95 M. avium clinical isolates were isolated from the sputum samples of patients with suspected tuberculosis between 2005 and 2012, which were collected from Fujian, Hunan, Jiangxi, Sichuan, and Anhui provinces in China. They were part of the routine hospital laboratory procedure. The species were identified via sequence analysis of hsp65, rpoB, and the 16S–23S ITS genes.13,14
Antimicrobial agents
Ofloxacin, ciprofloxacin, levofloxacin, moxifloxacin, sparfloxacin, streptomycin, amikacin, kanamycin, capreomycin, and tobramycin were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). All the antibiotics were freshly prepared before they were used.
Drug susceptibility tests
M. avium isolates were incubated in Difco Middlebrook 7H10 agar from BD (Franklin Lakes, NJ, USA) with 5% oleic acid–albumin–dextrose–catalase.12,15 The microdilution method was performed for fluoroquinolone and aminoglycoside susceptibility testing against M. avium using cation-adjusted Mueller-Hinton broth with the addition of 5% oleic acid–albumin–dextrose–catalase.12,15 The experiments were conducted in 96-well microplates. First, 0.5 McFarland standard bacterial suspensions were prepared. Second, the bacterial solutions and drug dilution mixtures were added to wells with a blank control included.12,15 Finally, the 96-well microplates were incubated at 37°C with 5% carbon dioxide. The minimum inhibitory concentration (MIC), MIC50, and MIC90 values for each antimicrobial agent were determined according to the previous method.12 Fluoroquinolone and aminoglycoside resistance were identified using the MIC breakpoints.12,16
PCR amplification and DNA sequencing
Genomic DNA was isolated from the M. avium isolates by heating the bacterial suspensions in Tris–EDTA buffer (pH =8.0) at 100°C for 30 min. The precipitant was removed by centrifugation at 12,000 rpm for 5 min.17 A 50-μL polymerase chain reaction (PCR) mixture, which included the following reagents, was then prepared as follows: 2 μL 2× PCR mixture, 1 μM forward primer, 1 μM reverse primer, and 5 μL of genomic DNA, as described previously.17 The gyrA, gyrB, and rpsL gene primers used in this study are listed in Table 1. PCR amplification was performed in a thermocycler with an initial denaturation step for 5 min at 94°C, followed by 35 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min, and a 5-min final extension at 72°C.17 The PCR products were sequenced at Tianyihuiyuan Bio Tech Co., Ltd. (Beijing, China). All the sequencing results were compared with the M. avium 104 complete genome sequences (NC_008595.1) in GenBank using the NCBI BLAST server (www.ncbi.nlm.nih.gov).
  | Table 1 The primers and sequence lengths for gyrA, gyrB, and rpsL genes Abbreviations: F, forward; R, reverse. |
Statistical analysis
The data obtained herein were analyzed using the SPSS v.17.0 software. The resistance percentages and the MIC50 and MIC90 values for the fluoroquinolone and aminoglycoside agents were calculated for the M. avium isolates.
Results
Antimicrobial resistance in the M. avium clinical isolates
The M. avium isolates displayed high resistance to ofloxacin (89/95, 93.68%) and ciprofloxacin (88/95, 92.63%). Moxifloxacin (18/95, 18.95%) showed the best activity in the strains tested. In terms of their MIC50 and MIC90 values, ofloxacin had the highest values and moxifloxacin had the lowest ones compared with the other fluoroquinolones. With the aminoglycosides, amikacin (2/95, 2.11%) demonstrated excellent activity against the isolates, whereas streptomycin (62/95, 65.26%) exhibited the worst activity. The MIC50 and MIC90 values of tobramycin were 1 and 16 μg/mL and kanamycin were 2 and >32 μg/mL, respectively (Table 2).
Mutations in gyrA and gyrB genes and fluoroquinolone resistance
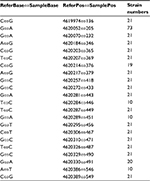
Fluoroquinolone resistance in the M. avium isolates was identified by screening for mutations in the gyrA and gyrB genes of this bacterium. Connections between fluoroquinolone resistance and gyrA and gyrB gene mutations were investigated. Among the 18 strains showing completely resistance to all the five fluoroquinolone agents, the most frequent SNPs were T→C (71/95, 74.74%), followed by A→G (64/95, 67.37%) and T→C (62/95, 65.26%) in the gyrA region of DNA gyrase (Table 3). The amino acid mutations occurred mainly at the sites of Gly2444Asp (GGT→GAT) (20/95, 21.05%), Ala2445Ser (GCC→TCC) (20/95, 21.05%), Ala2447Val (GCC→GTC) (20/95, 21.05%), Val2449Ile (GTC→ATC) (20/95, 21.05%), and Glu2450Gln (GAA→CAA) (20/95, 21.05%) (Table 4). Prominent SNPs in gyrB included A→C (69/95, 72.63%), C→T (51/95, 53.68%), and T→G (29/95, 30.53%) (Table 5), and their corresponding amino acid substitutions were Ile2160Val (ATT→GTT) (21/95, 22.11%), Ile2160Met (ATT→ATG) (20/95, 21.05%), and Ile2273Leu (ATC→CTC) (11/95, 11.58%) (Table 6).
Mutations in the rpsL gene and aminoglycoside resistance
Aminoglycoside resistance in M. avium isolates was identified from the mutation in the rpsL gene. Associations between aminoglycoside resistance in the isolates and rpsL gene mutation were focused on. Among the strains with aminoglycoside resistance, SNPs were detected mostly at position G→A (73/95, 76.84%) in rpsL. G→C (21/95, 22.11%) was commonly seen (Table 7). The amino acid mutations primarily involved Ala1539985Thr (GCC→ACC) (19/95, 20.00%), His1539992Asp (CAC→GAC) (19/95, 20.00%), and Gln1539983Glu (CAG→GAG) (18/95, 18.95%) (Table 8).
Discussion
In this study, the presence of SNPs associated with fluoroquinolone and aminoglycoside resistance in M. avium isolates was described. Among fluoroquinolones, ofloxacin was the worst activity and moxifloxacin was the best one against the M. avium isolates. Within aminoglycoside agents, streptomycin was the highest and capreomycin was the lowest resistance to the M. avium strains. Herein the SNPs, there were few reports about the M. avium species, but most in the M. tuberculosis.
From our assay, in gyrA, we found that the nonsynomymous mutations consisted of GCC→TCC (Ala→Ser) and GCC→GTC (Ala→Val), which were in agreement with the previous studies for M. tuberculosis.11,18–22 However, our findings were demonstrated that GTC→ATC (Val→Ile), GAA→CAA (Glu→Gln), and GGT→GAT (Gly→Asp) may be in the hotpot mutation region and crucially responsible for the fluoroquinolone resistance. Some studies indicated that the gyrA mutations of M. tuberculosis were in Ala90Thr, Asp94His, Asp94Val, Thr95Ser, Thr80Ala, and Ser91Pro, which were not found in our studies.23–28 According to the gyrB mutations, some studies were elucidated that there were the mutations in the Asn499Thr, Asp461Asn, and Gly512Arg.29–31 In our research, SNPs in the gyrB commonly included ATT→GTT (Ile→Val) and ATT→ATG (Ile→Met), which mutated in the 2160th position (it may be a hotpot mutation position) of the reference M. avium 104. Otherwise isoleucine might be the critical mutation amino acid.
Aminoglycoside resistance in mycobacteria usually occurs in the rpsL gene.32–34 In our current study, the SNPs were mostly described as G to A substitution in rpsL, followed by C to G substitution. The most common amino acid mutation mainly involved Ala→Thr and His→Asp. In previous studies, AAG→AGG (Lys43Arg) in rpsL was the most common mutation among the resistant M. tuberculosis isolates that showed an MDR phenotype, whereas an AAG→AGG (Lys88Arg) substitution was found to usually play a minor role and occur much less frequently than Lys43Arg.32,35–37
On the one hand, we found some isolates contained SNPs, but they were synonymous mutations. Some strains were resistant to fluoroquinolones and aminoglycosides in the absence of amino acid mutations, whose drug-resistant mechanism should be explored in the future. Furthermore, strains with higher drug resistance phenotypes had nonsynonymous mutations in gyrA and gyrB and rpsL genes. On the other hand, the isolates with synonymous mutations were resistant to drugs. There were 48 fluoroquinolone-resistant isolates with gyrA synonymous mutations and 52 resistant strains with gyrB synonymous mutations. In terms of aminoglycoside resistance, there were 12 drug-resistant strains with rpsL synonymous mutation.
It is known that drug resistance may be caused by mutations in the target regions of antimycobacterial drugs. The fluoroquinolone and aminoglycoside resistance regions need to be accurately identified in order to study the SNPs and amino acid mutations occurring in them. It also should be deeply researched whether DNA extracted from specimens may lack the resistant genotype and lead to a false low detection rate. Nevertheless, sequencing the regions of genes can be a convenient way to quickly identify antituberculosis drug resistance, thus enabling the selection of effective drugs for treatment of the disease.18,38
In addition to SNPs, there could be other drug resistance mechanisms, which should be discussed in the future. For one thing, the permeability in the cell wall to drugs and the existence of efflux pumps probably account for fluoroquinolone and aminoglycoside resistance in other resistant isolates that lack gyrA and gyrB and rpsL mutations. For another thing, fluoroquinolones can inhibit bacterial topoisomerase IV, which may inhibit DNA replication and transcription, and cause drug resistance.11 Furthermore, M. avium is known to have subspecies, such as M. avium subsp. avium, M. genavense, and M. avium subsp. paratuberculosis.29–31,39 These subspecies may differ in their drug resistance and SNP profiles, making them worthy of further research.
Conclusion
The fluoroquinolone and aminoglycoside resistance patterns and the SNP profiles of the clinical isolates of M. avium were considered. These data should be helpful for optimizing the treatment and preventing further transmission of drug-resistant M. avium strains. These findings have emphasized the need for the implementation of drug susceptibility along with accurate and rapid molecular tests for the detection of resistant mutations once M. avium is diagnosed.
Acknowledgments
We thank the staff of National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention. This study was financially supported by the Project of the Science and Technology of Shanxi Province for Youth Science Foundation (No 2016021161), the Science and Technology Innovation Team support project (No CX201412) from Changzhi Medical College, and innovation project of university students of Changzhi Medical College (No D2017015).
Disclosure
The authors report no conflicts of interest in this work.
References
Akram SM, Attia FN, editors. Mycobacterium avium intracellulare. StatPearls. Treasure Island (FL): StatPearls Publishing; 2017. | ||
Koh WJ. Nontuberculous mycobacteria – overview. Microbiol Spectr. 2017;5(1):1–7. | ||
Kang YA, Koh WJ. Antibiotic treatment for nontuberculous mycobacterial lung disease. Expert Rev Respir Med. 2016;10(5):557–568. | ||
Sommerstein R, Rüegg C, Kohler P, Bloemberg G, Kuster SP, Sax H. Transmission of Mycobacterium chimaera from heater-cooler units during cardiac surgery despite an ultraclean air ventilation system. Emerg Infect Dis. 2016;22(6):1008–1013. | ||
Boyle DP, Zembower TR, Reddy S, Qi C. Comparison of clinical features, virulence, and relapse among Mycobacterium avium complex species. Am J Respir Crit Care Med. 2015;191(11):1310–1317. | ||
Li G, Pang H, Guo Q, et al. Antimicrobial susceptibility and MIC distribution of 41 drugs against clinical isolates from China and reference strains of nontuberculous mycobacteria. Int J Antimicrob Agents. 2017;49(3):364–374. | ||
Avalos E, Catanzaro D, Catanzaro A, et al. Frequency and geographic distribution of gyrA and gyrB mutations associated with fluoroquinolone resistance in clinical Mycobacterium tuberculosis isolates: a systematic review. PLoS One. 2015;10(3):e012047012. | ||
Chaoui I, Oudghiri A, El Mzibri M. Characterization of gyrA and gyrB mutations associated with fluoroquinolone resistance in Mycobacterium tuberculosis isolates from Morocco. J Glob Antimicrob Resist. 2017;12:171–174. | ||
Hlaing YM, Tongtawe P, Tapchaisri P, et al. Mutations in streptomycin resistance genes and their relationship to streptomycin resistance and lineage of Mycobacterium tuberculosis Thai isolates. Tuberc Respir Dis. 2017;80(2):159–168. | ||
Rezaei F, Haeili M, Imani Fooladi A, Azari Garmjan GA, Feizabadi MM. Screening for streptomycin resistance conferring mutations in Mycobacterium tuberculosis isolates from Iran. J Chemother. 2017;29(1):14–18. | ||
Brown-Elliott BA, Nash KA, Wallace RJ Jr. Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria. Clin Microbiol Rev. 2012;25(3):545–582. | ||
Clinical and Laboratory Standards Institute. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes. Approved Standard. 2nd ed. Wayne: CLSI; 2011. Document M24-A2. | ||
Kim J-U, Cha C-H, An H-K. Multiplex real-time PCR assay and melting curve analysis for identifying Mycobacterium tuberculosis complex and nontuberculous mycobacteria. J Clin Microbiol. 2012;50(2):483–487. | ||
Pérez-Osorio AC, Boyle DS, Ingham ZK, et al. Rapid identifcation of mycobacteria and drug-resistant Mycobacterium tuberculosis by use of a single multiplex PCR and DNA sequencing. J Clin Microbiol. 2012;50(2):326–336. | ||
Hui P, Yi J, Kanglin W. Drug susceptibility of 33 reference strains of slowly growing mycobacteria to 19 antimicrobial agents. Biomed Res Int. 2017;2017:1584658. | ||
World Health Organization. Policy Guidance on Drug-Susceptibility Testing (DST) of Second-Line Antituberculosis Drugs. Geneva: WHO; 2008. | ||
Zhao X, Wang Y, Pang Y. Antimicrobial susceptibility and molecular characterization of Mycobacterium intracellulare in China. Infect Genet Evol. 2014;27:332–338. | ||
Xie YL, Chakravorty S, Armstrong DT, et al. Evaluation of a rapid molecular drug-susceptibility test for tuberculosis. N Engl J Med. 2017;377(11):1043–1054. | ||
Rindi L, Garzelli C. Genetic diversity and phylogeny of Mycobacterium avium. Infect Genet Evol. 2014;21:375–383. | ||
Karimi S, Mirhendi H, Zaniani FR, et al. Rapid detection of streptomycin-resistant Mycobacterium tuberculosis by rpsL restriction fragment length polymorphism. Adv Biomed Res. 2017;6:126. | ||
Bigi MM, Lopez B, Blanco FC, et al. Single nucleotide polymorphisms may explain the contrasting phenotypes of two variants of a multidrug-resistant Mycobacterium tuberculosis strain. Tuberculosis. 2017;103:28–36. | ||
Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31(17):2877–2878. | ||
Sandgren A, Strong M, Muthukrishnan P, et al. Tuberculosis drug resistance mutation database. PLoS Med. 2009;6(2):e1000002. | ||
Hoshide M, Qian L, Rodrigues C, et al. Geographical differences associated with single-nucleotide polymorphisms (SNPs) in nine gene targets among resistant clinical isolates of Mycobacterium tuberculosis. J Clin Microbiol. 2014;52(5):1322–1329. | ||
Martínez LM, Castro GP, Guerrero MI. A molecular platform for the diagnosis of multidrug-resistant and pre-extensively drug-resistant tuberculosis based on single nucleotide polymorphism mutations present in Colombian isolates of Mycobacterium tuberculosis. Mem Inst Oswaldo Cruz. 2016;111(2):93–100. | ||
Chien JY, Chiu WY, Chien ST, Chiang CJ, Yu CJ, Hsueh PR. Mutations in gyrA and gyrB among fluoroquinolone- and multidrug-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2016;60(4):2090–2096. | ||
Chen J, Peng P, Du Y, et al. Early detection of multidrug and pre-extensively drug-resistant tuberculosis from smear positive sputum by direct sequencing. BMC Infect Dis. 2017;17(1):300. | ||
Ajileye A, Alvarez N, Merker M, et al. Some synonymous and nonsynonymous gyrA mutations in Mycobacterium tuberculosis lead to systematic false-positive fluoroquinolone resistance results with the hain genoType MTBDRsl assays. Antimicrob Agents Chemother. 2017;61(4):e2169–e2116. | ||
Devasia R, Blackman A, Eden S, et al. High proportion of fluoroquinolone resistant Mycobacterium tuberculosis isolates with novel gyrase polymorphisms and a gyrA region associated with fluoroquinolone susceptibility. J Clin Microbiol. 2012;50(4):1390–1396. | ||
Willby M, Sikes RD, Malik S, Metchock B, Posey JE. Correlation between GyrA substitutions and ofloxacin, levofloxacin, and moxifloxacin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2015;59(9):5427–5434. | ||
Nosova EY, Bukatina AA, Isaeva YD, Makarova MV, Galkina KY, Moroz AM. Analysis of mutations in the gyrA and gyrB genes and their association with the resistance of Mycobacterium tuberculosis to levofloxacin, moxifloxacin and gatifloxacin. J Med Microbiol. 2013;62(Pt 1):108–113. | ||
Bernard C, Veziris N, Brossier F, et al. Molecular diagnosis of fluoroquinolone resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2015;59(3):1519–1524. | ||
Escribano I, Rodríguez JC, Llorca B, García-Pachon E, Ruiz M, Royo G. Importance of the efflux pump systems in the resistance of Mycobacterium tuberculosis to fluoroquinolones and linezolid. Chemotherapy. 2007;53(6):397–401. | ||
Yin X, Yu Z. Mutation characterization of gyrA and gyrB genes in levofloxacin-resistant Mycobacterium tuberculosis clinical isolates from Guangdong Province in China. J Infect. 2010;61(2):150–154. | ||
Jagielski T, Ignatowska H, Bakuła Z, et al. Screening for streptomycin resistance-conferring mutations in Mycobacterium tuberculosis clinical isolates from Poland. PLoS One. 2014;9(6):e100078. | ||
Bauskenieks M, Pole I, Skenders G, et al. Genotypic and phenotypic characteristics of aminoglycoside-resistant Mycobacterium tuberculosis isolates in Latvia. Diagn Microbiol Infect Dis. 2015;81(3):177–182. | ||
Sun H, Zhang C, Xiang L, et al. Characterization of mutations in streptomycin-resistant Mycobacterium tuberculosis isolates in Sichuan, China and the association between Beijing-lineage and dual-mutation in gidB. Tuberculosis. 2016;96:102–106. | ||
Zhang H, Li D, Zhao L, et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet. 2013;45(10):1255–1260. | ||
Lee MR, Chien JY, Huang YT, et al. Clinical features of patients with bacteraemia caused by Mycobacterium avium complex species and antimicrobial susceptibility of the isolates at a medical centre in Taiwan, 2008-2014. Int J Antimicrob Agents. 2017;50(1):35–40. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.