Back to Journals » Cancer Management and Research » Volume 11
Silencing Of hsa_circ_0008450 Represses Hepatocellular Carcinoma Progression Through Regulation Of microRNA-214-3p/EZH2 Axis
Authors Lin T , Dai Y, Guo X, Chen W, Zhao J, Cao L, Wu Z
Received 10 July 2019
Accepted for publication 9 October 2019
Published 24 October 2019 Volume 2019:11 Pages 9133—9143
DOI https://doi.org/10.2147/CMAR.S222716
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Beicheng Sun
Tianyu Lin,1,* Yi Dai,1,* Xinli Guo,2 Wenchao Chen,1 Jiangang Zhao,3 Liping Cao,1 Zhengrong Wu1
1Department of General Surgery, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou 310000, People’s Republic of China; 2Department of Operating Room, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou 310000, People’s Republic of China; 3Department of General, Visceral and Tumor Surgery, University Hospital Cologne, Cologne 50934, Germany
*These authors contributed equally to this work
Correspondence: Liping Cao; Zhengrong Wu
Department of General Surgery, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, No. 3, Qingchun Road, Hangzhou 310000, People’s Republic of China
Tel +86-571-86006961
Fax +86-571-86006961
Email [email protected]; [email protected]
Purpose: Circular RNA (circRNA) hsa_circ_0008450 has been shown to be up-regulated in hepatocellular carcinoma (HCC). However, the functional role of hsa_circ_0008450 and its molecular mechanism are still unknown.
Patients and methods: We used qRT-PCR and Western blot to examine the expression levels of hsa_circ_0008450, microRNA-214-3p (miR-214-3p), and enhancer of zeste homolog 2 (EZH2) protein. CCK8 assay and wound healing assay were used to detect cell viability and cell migration capability. Cell apoptosis was assessed by flow cytometry. Luciferase reporter assay was used to explore the interaction among hsa_circ_0008450, miR-214-3p, and EZH2.
Results: hsa_circ_0008450 was significantly increased in HCC tissues and cells. Furthermore, knockdown of hsa_circ_0008450 in HCC cells inhibited cell proliferation, invasion, and migration. Mechanically, hsa_circ_0008450 promoted the expression of EZH2 protein through sponging miR-214-3p. Knockdown of circ_0008450 suppressed tumorigenesis of HCC cells in vivo.
Conclusion: Knockdown of hsa_circ_0008450 inhibits HCC progression by regulating miR-214-3p/EZH2 axis. This study suggests that hsa_circ_0008450 may serve as a novel target for the treatment of HCC.
Keywords: circular RNAs, hepatocellular carcinoma, hsa_circ_0008450, miR-214-3p, enhancer of zeste homolog 2
Introduction
Regarded as the most common histologic type of primary liver cancer, hepatocellular carcinoma (HCC) is the third leading cause of cancer-related mortalities.1–3 For the moment, surgical resection, liver transplantation, systemic therapy, and chemoradiotherapy (doxorubicin and cisplatin) are the main options for HCC treatment.4 Although substantial progress has been made in surgical techniques and postoperative chemotherapy in recent years, the clinical outcome of HCC patients is still poor. Improvement in five-year survival rate is not satisfactory, largely due to recurrence and metastasis.4 Thus, it is of great significance to explore the pathogenesis of HCC and find new therapeutic targets to design more effective treatment methods.
Circular RNAs (circRNAs) are an important class of endogenous non-coding RNAs that are widely exist in mammalian cells.5,6 CircRNAs are featured with a covalently closed RNA molecules in loop structures with neither 5ʹ cap nor 3ʹ polyadenylated tail.7,8 CircRNAs have been discovered to regulate the cell biological behaviors in cancer cells, such as cell proliferation, apoptosis, cancer migration, and invasiveness.9,10 Previous studies have shown that circRNAs participate in carcinogenesis of several cancers, such as ovarian cancer,11 gastric cancer,12 and colorectal cancer.13 Recently, hsa_circ_0008450, as a novel circRNA, has been found to be upregulated in HCC tissues by high-throughput circRNA sequencing.14 Zhang et al found that hsa_circ_0008450 was up-regulated in HCC tissues and cells. Silencing of hsa_circ_0008450 inhibited HCC cell growth, invasion, and migration.15
MicroRNAs (miRNAs), a class of endogenous short noncoding RNAs, mediate the post-transcriptional negative regulation of expression of target genes.16 In recent years, the oncogenic or tumor-suppressive function of miRNAs has been reported.17,18 Dysregulation of miR-214-3p has been identified in HCC.19 It has been reported that the expression of miR-214-3p was down-regulated in HCC tissues. Moreover, recent studies have demonstrated that miR-214-3p suppressed HCC cell proliferation and enhanced cell apoptosis by directly down-regulating MELK expression.20
Enhancer of zeste 2 homolog (EZH2) is the catalytic subunit of the polycomb repressive complex 2, which is a complex that methylates lysine 27 of histone H3 (H3K27) to repress its gene expression.21–23 Meanwhile, EZH2 has been well studied in tumor progression, such as breast cancer,24 prostate cancer,25 and colorectal cancer.26 Intriguingly, EZH2 was also reported to be a critical regulator of circRNA in cancers.27 However, the interaction between hsa_circ_0008450 and miR-214-3p/EZH2 in HCC has not been reported yet.
In this study, we determined that hsa_circ_0008450 was significantly up-regulated in HCC tissues and cells. In addition, hsa_circ_0008450 regulated EZH2 expression by acting as a miR-214-3p sponge. These findings indicated that down-regulation of hsa_circ_0008450 may be a potential therapeutic strategy for HCC.
Materials And Methods
Patient Tissue Samples
Thirty pairs of HCC tissues and adjacent normal tissues were obtained at Department of General Surgery, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University. Tissue samples were immediately plunged into liquid nitrogen and stored at ‒80°C for further use. Clinicopathological features of patients with HCC were shown in Table 1. Prior written informed consent was obtained from every patient, and all of the experimental protocols of the study were approved by Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University.
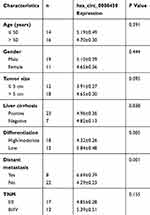 |
Table 1 The Correlation Between hsa_circ_0008450 Expression And Clinicopathologic Features Of HCC Patients (n=30) |
Cell Lines And Cell Culture
Human normal liver cell line (L02) and the human HCC cell lines (SMMC7721, Sk-Hep-1, HepG2, Huh-7, and HCCLM3) were purchased from the American Tissue Culture Collection (ATCC, Manassas, VA, USA). They were cultured in Roswell Park Memorial Institute-1640 medium (RPMI-1640; HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS, Gibco, USA) in a humidified atmosphere with 5% v/v CO2 at 37°C.
Cell Transfection
Negative control miRNA (miR-NC), miR-214-3p mimics, miR-214-3p inhibitor, small interfering RNA targeting hsa_circ_0008450 (si-circ_0008450) or EZH2, and siRNA negative control (si-NC) were purchased from RiboBio (Shanghai, China). Huh-7 and HCCLM3 cells were seeded in 6-well plates and cultured for 24 h, and then cells were co-transfected with 50 nM siRNAs or si-NC and miR-NC or miR-214-3p mimics or miR-214-3p inhibitor by using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. The knockdown efficiency of si-circ_0008450 was determined by quantitative real-time polymerase chain reaction (qRT-PCR).
RNA Extraction And qRT-PCR Assay
Total RNAs were isolated from HCC tissues, adjacent normal tissues and cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Total RNA (1 μg) was subjected to reverse transcription using PrimeScript RT Reagent Kit (Takara, Tokyo, Japan). qRT-PCR was performed with SYBR green (Takara) by Bio-Rad CFX96 system (Bio-Rad, Hercules, CA, USA) to determine the expression of hsa_circ_0008450, miR-214-3p, and EZH2 mRNA. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 were used as internal controls. The expression levels of genes were calculated by the 2−ΔΔCt method.
Dual-Luciferase Reporter Assay
The online prediction databases including Starbase v3.0 (http://starbase.sysu.edu.cn/) and Targetscan Release 7.2 (http://www.targetscan.org/vert_72/) were used to predict the potential miRNA target of hsa_circ_0008450 and downstream targets of miR-214-3p, respectively. The sequences of hsa_circ_0008450 and EZH2 were amplified by RT-PCR and cloned into the luciferase vector psiCHECK-2 (Promega, Madison, WI, USA). Recombinant mutant-type (MUT) of hsa_circ_0008450 and EZH2 were also established. For luciferase assays, Huh-7 and HCCLM3 cells (1×105) were seeded into 24-wells and co-transfected with luciferase reporter vectors containing the WT or MUT hsa_circ_0008450 (0.5 μg) or EZH2 (0.5 μg) and miR-214-3p mimics or negative control (50 nM) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientifc, Inc.). The luciferase activity was measured using a dual luciferase reporter assay kit (Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer’s protocol.
Cell Proliferation Assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Kumamoto, Japan) was performed to test Huh-7 and HCCLM3 cell proliferation. In brief, Huh-7 and HCCLM3 cells were seeded in 96-well plates at a density of 1×104 cells/well and cultured for indicated times at 37°C with 5% v/v CO2. Then, the cells were treated with 10 µl CCK-8 solution at 37°C for 2 h. Subsequently, the absorbance was assessed by spectrophotometer at 450 nm.
Wound Healing Assay
Wound-healing assay was used to measure the ability of cell migration. Huh-7 and HCCLM3 cells were seeded in 6-well plates (1×105 cells/well) and cultured for 24 h. One scratch was generated by a pipette tip when a single layer of cells was formed. Then, the cells were gently washed with sterilized phosphate buffer saline (PBS). Subsequently, serum-free medium was added into the wells. Images were captured at 0 h and 24 h under a microscope with a digital camera system (Olympus, Tokyo, Japan). Scratch width between the two linear regions was quantitated for assessing capacity of cell migration. The migration rate was calculated using the following formula: (width of the wound width at 0 h ‒ the wound width at 24 h)/the wound width at 0 h.
Flow Cytometry Assay
The apoptosis of HCC cells was detected with Annexin V-FITC Apoptosis Detection Kit (Beyotime Biotechnology Co. Ltd., Shanghai, China). Briefly, tumor cells were cultured in 6-well plates at a density of 4×105 cells/well. Subsequently, the transfected Huh-7 and HCCLM3 cells were collected and washed with 1×PBS. And the cell lines were added with 5μl of Annexin V-FITC and 5μl of propidium iodide (PI) and cultured for 10 mins at room temperature in the dark. Then, the apoptosis of the cells were measured by fluorescence activated cell sorting (FACS).
Western Blot
Treated Huh-7 and HCCLM3 cells were lysed in RIPA buffer (Beyotime Biotechnology, Dalian, China). Proteins were separated on dodecyl sulfate, sodium salt (SDS)-Polyacrylamide gel electrophoresis (PAGE) and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA). After blocking, the membranes were incubated with the primary antibodies (EZH2: Abcam, Cambridge, MA; GAPDH: Abcam) overnight at 4°C followed by incubation with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies at room temperature for 2 h. Finally, the protein expression level was normalized to GAPDH and quantified using the Image J software (National Institutes of Health, Bethesda, MD).
Nude Mouse Xenograft Model
One-month-old male BALB/c nude mice were purchased from Slac Laboratory Animal Center (Shanghai, China). The mice were randomly divided into 2 groups and maintained in the animal care facility under specific pathogen-free conditions. Next, the Huh-7 cells (2×106) transfected with si-NC and si-circ_0008450 were subcutaneously injected into the unde mice. Then, the length and width of the tumor were measured every one week to calculate the tumor volumes by the formula: volume = 0.5 × length × width2. The mice were sacrificed after 5 weeks and the tumors were removed and weighed. All animal experiments were approved by the Animal Care and Use Committee of Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University.
Statistical Analyses
Data from at least three independent experiments were expressed as mean ± standard deviation (SD). The difference between groups was analyzed using one-way ANOVA or Student’s t-test by SPSS 20.0 (IBM, NY, USA) software. A P-value of less than 0.05 was considered statistically significant.
Results
hsa_circ_0008450 Is Up-Regulated In HCC Tissue And Cell Lines
To validate the ectopic expression of hsa_circ_0008450 in HCC, qRT-PCR was performed in 30 pairs of HCC tumor tissues and adjacent normal tissues. The results showed that the expression level of hsa_circ_0008450 was significantly higher in HCC tissues (Figure 1A) than that in adjacent normal tissues. Then, we examined hsa_circ_0008450 expression through qRT-PCR in L02 cells and HCC cell lines (SMMC7721, Sk-Hep-1, HepG2, Huh-7, and HCCLM3). The hsa_circ_0008450 expression was up-regulated in HCC cell lines (Figure 1B). Therefore, these results suggested that hsa_circ_0008450 may be involved in HCC progression.
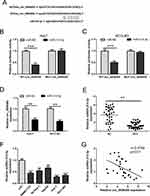
hsa_circ_0008450 Acts As A miRNA Sponge To Regulate miR-214-3p
To make clear the potential molecular mechanisms of hsa_circ_0008450 in HCC progression, we used Starbase3 to predict its targeted miRNA. The predicted result showed that hsa_circ_0008450 sequence contained miR-214-3p binding sites (Figure 2A). Then we performed luciferase reporter assays to confirm this possibility. The results indicated that the luciferase activity was markedly suppressed by miR-214-3p mimics (Figure 2B and C) in Huh-7 and HCCLM3 cells co-transfected with WT-circ_0008450 vector and miR-214-3p mimics. By contrast, the luciferase activity was un-affected in cells co-transfected with MUT-circ_0008450 vector and miR-214-3p mimics (Figure 2B and 2C). Furthermore, upregulation of miR-214-3p markedly decreased the expression level of hsa_circ_0008450 in Huh-7 and HCCLM3 cells (Figure 2D). Moreover, qRT-PCR was performed to detect miR-214-3p expression in HCC tumor tissues and cell lines. The expression of miR-214-3p was down-regulated in HCC tissue and cells (Figure 2E and F). Pearson’s correlation analysis showed that the expression of miR-214-3p was negatively related to hsa_circ_0008450 expression in HCC tumor tissues (Figure 2G). Therefore, these data indicated that hsa_circ_0008450 acted as a miRNA sponge to regulate miR-214-3p.
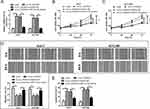
hsa_circ_0008450 Knockdown Represses Proliferation, Induced Apoptosis, And Reduced Migration In HCC Cells Via Regulating miR-214-3p
The above results showed that hsa_circ_0008450 served as a sponge of miR-214-3p in HCC cells. Thus, it is essential to explore whether hsa_circ_0008450 could regulate HCC progression by sponging miR-214-3p. The Huh-7 and HCCLM3 cells were transfected with si-circ_0008450 or co-transfected with si-circ_0008450 and miR-214-3p inhibitor. The result of qRT-PCR analysis showed that miR-214-3p expression in the si-circ_0008450 group was significantly up-regulated compared with the si-NC group, but this action was abolished by transfection of miR-214-3p inhibitor (Figure 3A). CCK-8 assay and wound healing assay indicated that hsa_circ_0008450 knockdown could markedly decrease the proliferation and migration abilities of Huh-7 and HCCLM3 cells. However, the reduction of cell proliferation and migration abilities was attenuated after co-transfection with si-circ_0008450 and miR-214-3p inhibitor (Figure 3B–D). Moreover, flow cytometry analysis was performed to further evaluate the effect of hsa_circ_0008450 knockdown on cell apoptosis in HCC cell lines. As shown in Figure 3E, hsa_circ_0008450 knockdown promoted the apoptosis of HCC cells, which was reversed by miR-214-3p inhibitor. These findings suggested that hsa_circ_0008450 knockdown could inhibit cell proliferation, migration, and promoted apoptosis by enhancing miR-214-3p expression in HCC cells.
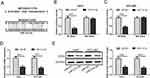
EZH2 Is A Direct Target Gene Of miR-214-3p
To confirm whether EZH2 was a direct target of miR-214-3p, the online predict software Targetscan was used to identify the potential binding sites. We found the 3ʹ-untranslated region (3ʹUTR) of EZH2 mRNA contained the potential binding sites of miR-214-3p (Figure 4A). Next we performed dual luciferase reporter assay to confirm this possibility. Luciferase reporter plasmids containing WT-EZH2 or MUT-EZH2 were transfected into Huh-7 and HCCLM3 cells together with miR-214-3p or miR-NC. In the reporter assay, luciferase activity was markedly inhibited by miR-214-3p mimics in Huh-7 and HCCLM3 cells transfected with WT-EZH2 plasmids. While mutations of the predictive miR-214-3p binding sites in EZH2 abolished this effect (Figure 4B and C). Furthermore, the qRT-PCR and Western blot showed that the mRNA and protein expression levels of EZH2 were significantly decreased in HCC cells transfected with miR-214-3p mimics (Figure 4D and E). These results supported that EZH2 was a direct target gene of miR-214-3p.
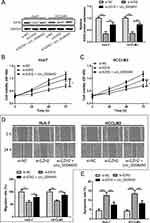
hsa_circ_0008450 Attenuates The Effect Of EZH2 Knockdown On HCC Cell Progression
To determine the relationship between hsa_circ_0008450 and EZH2 in HCC cells, Huh-7 and HCCLM3 cells were co-transfected with si-EZH2 and hsa_circ_0008450. As shown in Figure 5A, the protein expression of EZH2 was down-regulated in Huh-7 and HCCLM3 cells when transfected with si-EZH2 plasmid. EZH2 knockdown-induced repression of EZH2 protein expression was attenuated by upregulation of hsa_circ_0008450 (Figure 5A). EZH2 knockdown markedly decreased the proliferation and migration abilities of HCC cells, but these inhibitory effects were attenuated by upregulation of hsa_circ_0008450 (Figure 5B–E). Flow cytometry analysis showed that EZH2 knockdown promoted the apoptosis of HCC cells. These data indicated that hsa_circ_0008450 attenuated the effect of EZH2 knockdown on HCC cell progression.
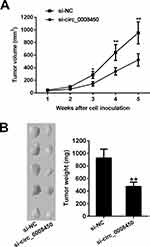
hsa_circ_0008450 Knockdown Inhibits Tumor Growth In Vivo
To further investigate the biological function of hsa_circ_0008450 in HCC, we performed the xenograft tumor experiment in nude mice. We found that tumor volumes and weights in the si-circ_0008450 group were significantly less than those in the si-NC group (Figure 6A and B). Thus, these results showed that circ_0008450 knockdown suppressed tumorigenesis of HCC cells in vivo.
Discussion
CircRNAs have been reported as novel biomarkers in cancer diagnosis and prognosis.28,29 For example, high expression of circ_0006528 was closely associated with tumor-node-metastasis growth and poor prognosis.30 Hsa_circ_0008450 has been found to be increased in HCC tissues compared with the corresponding normal tissue samples by high-throughput circRNA sequencing and qRT-PCR.14,15 Our results also showed that the expression of hsa_circ_0008450 was remarkably up-regulated in HCC tissues and cells, and our findings suggested that hsa_circ_0008450 might acted as an oncogene to promote HCC progression. These data implied that hsa_circ_0008450 might be a potential biomarker for HCC diagnosis and prognosis.
The elucidation of potential molecular mechanisms of hsa_circ_0008450 was helpful to develop effective and reasonable therapies of HCC. In our study, hsa_circ_0008450 sponged miR-214-3p to accelerate the proliferation and migration of HCC cells. Apart from our findings, more and more evidences suggested that circRNAs can function as miRNA sponges in the progression of tumors, serving as tumor suppressor genes.31 Recently, it has been reported that hsa_circ_0008450 acted as a sponge of miR-548p to promote the occurrence and development of HCC.15 Furthermore, circRNA_33287 acted as a sponge of miR-214-3p to promote the osteogenic differentiation.32 Our study demonstrated that hsa_circ_0008450 regulated the HCC cell proliferation, migration, and apoptosis and inhibited miR-214-3p expression. Thus, the oncogenic function of hsa_circ_0008450 might be attributed to its suppression on miR-214-3p.
Previously, EZH2 has been found to be involved in the development of human cancers.33–35 The precise mechanism by which EZH2 regulated proliferation, apoptosis, and migration of cancer cells remains unclear. Accumulating studies demonstrated that miRNA might produce tumor suppressors by regulating the expression of EZH2.36,37 For example, miR-124 directly targeted the 3ʹUTR of EZH2 mRNA to inhibit the HCC cell invasion and metastasis.36 Cui et al have found that miR-137 inhibited HCC growth and metastasis by suppressing the expression of EZH2.38 Here, bioinformatics analysis and dual luciferase reporter assay showed that EZH2 was a target gene of miR-214-3p, and we identified the functional role of miR214-3p/EZH2 axis in HCC cells.
Meanwhile, circRNA/miRNA/mRNA axis has been studied in onset and progression of cancer.39,40 For instance, hsa_circ_100395, as an oncogene, inhibited lung cancer malignant behaviors via modulating miR-1228/TCF21 axis.41 Moreover, hsa_circ_0071589 acted as a sponge of miR-600 to regulate the expression of EZH2, thus suppressing colorectal cancer progression.26 Furthermore, Zou et al discovered that hsa_circ_0101432 promoted HCC progression via inhibiting miR-1258 and miR-622 and increasing MAPK1 mRNA expression.42 In the present study, further functional experimental results showed that up-regulation of hsa_circ_0008450 attenuated the inhibitory effects of EZH2 knockdown on aggressive biological behaviors of HCC cells. All these data demonstrated that hsa_circ_0008450 acted as an oncogene to promote HCC malignant progression via modulating miR-214-3p/EZH2 axis.
Conclusion
In conclusion, our work identified that hsa_circ_0008450 expression level was increased in HCC tissues and cells. Knockdown of hsa_circ_0008450 suppressed aggressive biological behaviors of HCC cells via modulating the miR-214-3p/EZH2 axis. Better understanding of the roles of circRNA during HCC initiation and progression will facilitate the development of therapeutics against human HCC.
Abbreviations
circRNA, Circular RNA; HCC, hepatocellular carcinoma; EZH2, enhancer of zeste homolog 2; miRNAs, MicroRNAs; H3K27, methylate lysine 27 of histone H3; qRT-PCR, quantitative real-time polymerase chain reaction; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; MUT, mutant-type; PBS, phosphate buffer saline; PI, propidium iodide; FACS, fluorescence activated cell sorting; PAGE, Polyacrylamide gel electrophoresis; HRP, horseradish peroxidase; SD, standard deviation.
Acknowledgment
This work was funded by the Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (No. 2018246436).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lafaro KJ, Demirjian AN, Pawlik TM. Epidemiology of hepatocellular carcinoma. Surg Oncol Clin N Am. 2015;24(1):1–17. doi:10.1016/j.soc.2014.09.001
2. McGlynn KA, Petrick JL, London WT. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clin Liver Dis. 2015;19(2):223–238. doi:10.1016/j.cld.2015.01.001
3. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi:10.3322/caac.21349
4. Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, Kamel IR, Ghasebeh MA, Pawlik TM. Hepatocellular carcinoma: from diagnosis to treatment. Surg Oncol. 2016;25(2):74–85. doi:10.1016/j.suronc.2016.03.002
5. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi:10.1038/nbt.2890
6. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
7. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20(12):1829–1842. doi:10.1261/rna.047126.114
8. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
9. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310.
10. Hu C, Wang Y, Li A, Zhang J, Xue F, Zhu L. Overexpressed circ_0067934 acts as an oncogene to facilitate cervical cancer progression via the miR-545/EIF3C axis. J Cell Physiol. 2019;234(6):9225–9232. doi:10.1002/jcp.27601
11. Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi:10.1038/srep08057
12. Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med.2017;6(6):1173–1180.
13. Zhu M, Xu Y, Chen Y, Yan F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–144. doi:10.1016/j.biopha.2016.12.097
14. Cai H, Hu B, Ji L, Ruan X, Zheng Z. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway. Am J Transl Res. 2018;10(6):1690–1702.
15. Zhang J, Chang Y, Xu L, Qin L. Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p. J Cell Biochem. 2019;120(6):9487–9494. doi:10.1002/jcb.v120.6
16. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi:10.1146/annurev-biochem-060308-103103
17. Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223(2):102–115. doi:10.1002/path.2806
18. Gentilin E, Degli Uberti E, Zatelli MC. Strategies to use microRNAs as therapeutic targets. Best Pract Res Clin Endocrinol Metab. 2016;30(5):629–639. doi:10.1016/j.beem.2016.10.002
19. Shi KQ, Lin Z, Chen XJ, et al. Hepatocellular carcinoma associated microRNA expression signature: integrated bioinformatics analysis, experimental validation and clinical significance. Oncotarget. 2015;6(28):25093–25108. doi:10.18632/oncotarget.4437
20. Li Y, Li Y, Chen Y, et al. Correction to: microRNA-214-3p inhibits proliferation and cell cycle progression by targeting MELK in hepatocellular carcinoma and correlates cancer prognosis. Cancer Cell Int. 2018;18:55.
21. Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi:10.1038/nature09784
22. McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi:10.1038/nature11606
23. Donaldson-Collier MC, Sungalee S, Zufferey M. EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nat Genet. 2019;51(3):517–528. doi:10.1038/s41588-018-0338-y
24. Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100(20):11606–11611. doi:10.1073/pnas.1933744100
25. Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi:10.1038/nature01075
26. Yong W, Zhuoqi X, Baocheng W, Dongsheng Z, Chuan Z, Yueming S. Hsa_circ_0071589 promotes carcinogenesis via the miR-600/EZH2 axis in colorectal cancer. Biomed Pharmacother. 2018;102:1188–1194. doi:10.1016/j.biopha.2018.03.085
27. Xue J, Liu Y, Luo F, et al. Circ100284, via miR-217 regulation of EZH2, is involved in the arsenite-accelerated cell cycle of human keratinocytes in carcinogenesis. Biochim Biophys Acta Mol Basis Dis. 2017;1863(3):753–763.
28. Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomarkers. 2016;16(1):161–169.
29. Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine. 2016;95(22):e3811. doi:10.1097/MD.0000000000003811
30. Gao D, Qi X, Zhang X, Fang K, Guo Z, Li L. hsa_circRNA_0006528 as a competing endogenous RNA promotes human breast cancer progression by sponging miR-7-5p and activating the MAPK/ERK signaling pathway. Mol Carcinog. 2019;58(4):554–564. doi:10.1002/mc.v58.4
31. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi:10.1002/hep.29270
32. Peng W, Zhu S, Chen J, Wang J, Rong Q, Chen S. Hsa_circRNA_33287 promotes the osteogenic differentiation of maxillary sinus membrane stem cells via miR-214-3p/Runx3. Biomed Pharmacother. 2019;109:1709–1717.
33. Wassef M, Luscan A, Aflaki S, et al. EZH1/2 function mostly within canonical PRC2 and exhibit proliferation-dependent redundancy that shapes mutational signatures in cancer. Proc National Academy Sci USA. 2019;116(13):6075–6080. doi:10.1073/pnas.1814634116
34. Yamagishi M, Uchimaru K. Targeting EZH2 in cancer therapy. Curr Opin Oncol. 2017;29(5):375–381. doi:10.1097/CCO.0000000000000390
35. Yan KS, Lin CY, Liao TW, et al. EZH2 in cancer progression and potential application in cancer therapy: a friend or foe? Int J Mol Sci. 2017;18:6.
36. Zheng F, Liao YJ, Cai MY, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61(2):278–289. doi:10.1136/gut.2011.239145
37. Tian Z, Li Z, Zhu Y, et al. Hypermethylation-mediated inactivation of miR-124 predicts poor prognosis and promotes tumor growth at least partially through targeting EZH2/H3K27me3 in ESCC. Clin Exp Metastasis. 2019;36:381–391. doi:10.1007/s10585-019-09974-1
38. Cui S, Sun Y, Liu Y, et al. MicroRNA-137 has a suppressive role in liver cancer via targeting EZH2. Mol Med Rep. 2018;17(5):7460.
39. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317.
40. Deng N, Li L, Gao J, et al. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun. 2018;495(1):189–196. doi:10.1016/j.bbrc.2017.11.028
41. Chen D, Ma W, Ke Z, Xie F. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17(16):2080–2090.
42. Zou H, Xu X, Luo L, et al. Hsa_circ_0101432 promotes the development of hepatocellular carcinoma (HCC) by adsorbing miR-1258 and miR-622. Cell Cycle. 2019;18(19):2398–2413.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.