Back to Journals » Cancer Management and Research » Volume 13
Silencing of circHIPK3 Sensitizes Paclitaxel-Resistant Breast Cancer Cells to Chemotherapy by Regulating HK2 Through Targeting miR-1286
Authors Ni J, Xi X, Xiao S, Xiao X
Received 20 February 2021
Accepted for publication 11 June 2021
Published 12 July 2021 Volume 2021:13 Pages 5573—5585
DOI https://doi.org/10.2147/CMAR.S307595
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Seema Singh
Jun Ni, Xun Xi, Sujian Xiao, Xigang Xiao
Department of Breast and Thyroid Surgery, People’s Hospital of Ganzhou City, Ganzhou, Jiangxi, People’s Republic of China
Correspondence: Xigang Xiao
Department of Breast and Thyroid Surgery, People’s Hospital of Ganzhou City, No. 16 Meiguan Road, Ganzhou City, Jiangxi Province, 341000, People’s Republic of China
Tel/Fax +86-797-5889738
Email [email protected]
Background: Resistance development to paclitaxel (PTX) has become a major obstacle in the successful treatment of breast cancer (BC). Circular RNAs (circRNAs) have been identified as essential regulators in PTX resistance of BC. Here, we explored the precise roles of circRNA homeodomain interacting protein kinase 3 (circHIPK3, circ_0000284) in PTX resistance of BC.
Methods: The expression levels of circHIPK3, microRNA (miR)-1286, and hexokinase 2 (HK2) were detected by quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot. Ribonuclease R (RNase R) assay was used to confirm the stability of circHIPK3. Cellular localization of circHIPK3 was assessed by subcellular localization assay. The half maximal inhibitory concentration (IC50) value for PTX was measured by Cell Counting Kit-8 (CCK-8) assay. Cell colony formation, cell cycle distribution, and apoptosis were gauged by colony formation assay and flow cytometry, respectively. Animal studies were performed to evaluate the role of circHIPK3 in vivo. The direct relationship between miR-1286 and circHIPK3 or HK2 was verified by dual-luciferase reporter and RNA immunoprecipitation (RIP) assays.
Results: Our results showed that circHIPK3 was up-regulated in PTX-resistant BC tissues and cells compared with the sensitive counterparts. The silencing of circHIPK3 promoted PTX sensitivity of PTX-resistant BC cells in vitro and in vivo. CircHIPK3 directly targeted miR-1286, and miR-1286 acted as a downstream mediator of circHIPK3 function in vitro. HK2 was a direct target of miR-1286, and circHIPK3 modulated HK2 expression through miR-1286. The increased expression of miR-1286 sensitized PTX-resistant BC cells to PTX in vitro by down-regulating HK2.
Conclusion: Our findings demonstrated that the silencing of circHIPK3 sensitized PTX-resistant BC cells to PTX therapy at least in part via the regulation of the miR-1286/HK2 axis.
Keywords: chemoresistance, circHIPK3, miR-1286, HK2
Introduction
Breast cancer (BC) remains the most common type of cancer among women.1 Paclitaxel (PTX), a type of chemotherapeutic drug taxanes, is an antineoplastic drug, which induces cell death by impacting microtubule stabilization.2 PTX has been clinically approved and used for BC treatment and is effective against early and metastatic BC.3 Unfortunately, resistance development to PTX treatment has become a major problem in successful BC treatment.4 Understanding the molecular basis of PTX resistance is indispensable for improving BC therapies.
Covalently closed circular RNAs (circRNAs) are naturally occurring RNA circles that have no free 3ʹ and 5ʹ ends.5 Many circRNAs involve in post-transcriptional regulation by inhibiting microRNA (miRNA) activity.6 CircRNAs have been identified as essential regulators in the chemoresistance development of various cancers, including BC.7 For instance, the overexpression of circ_0005728 contributed to doxorubicin resistance in BC through targeting miR-512-3p.8 The up-regulation of circ_0001982 induced the development of PTX resistance of BC by the regulation of miR-140-5p.9 As for circRNA homeodomain interacting protein kinase 3 (circHIPK3, circ_0000284), produced by the back-splicing of HIPK3 mRNA, it has been identified as an oncogenic driver in various cancers,10 including BC.11 Importantly, recent evidence highlighted that circHIPK3 could enhance the chemoresistance development of pancreatic cancer and colorectal cancer.12,13 However, the critical role of circHIPK3 in PTX resistance of BC remains to be elucidated.
MiRNAs are attractive candidates as upstream modulators of the chemoresistance development because they can control entire sets of genes.14,15 Examples of miRNAs suppressing BC chemoresistance included miR-708-3p and miR-7, which inhibited the translation of their targeted mRNAs.16,17 Conversely, exosome-mediated miR-155 enhanced the chemoresistance development in BC.18 When we used the online algorithm to identify the targeted miRNAs of circHIPK3, we found that circHIPK3 harbored a putative complementary site for miR-1286, an anti-tumor miRNA in osteosarcoma.19 However, it is still unexplored whether the regulation of circHIPK3 on BC PTX resistance is mediated by miR-1286.
Hexokinase 2 (HK2), a pivotal enzyme of glucose metabolism, is expressed at a high level in cancer cells and is widely recognized as an oncogenic driver in various cancers, including BC.20–23 Moreover, HK2 is reported to be implicated in the pathogenesis of BC chemoresistance.24,25 Here, we set to elucidate the precise role of circHIPK3 in PTX resistance of BC. Our results showed that the silencing of circHIPK3 sensitized PTX-resistant BC cells to PTX therapy by the regulation of the miR-1286/HK2 axis.
Materials and Methods
Clinical Specimens
The clinical specimens of 76 cases, including 13 cancer tissue samples from primary BC patients (defined as PTX-sensitive tissues), 25 cancer tissue samples from recurrent patients after treatment based on PTX chemotherapy (defined as PTX-resistant tissues), and 38 adjacent normal breast tissue samples from these patients were obtained from People’s Hospital of Ganzhou City. None of these patients received chemotherapy or radiotherapy before resection. All specimens were collected after surgical resection and only used to evaluate the expression of circHIPK3, miR-1286, and HK2 because of the small sample size and experimental requirements. Ethics approval for this project was obtained from the Human Research Ethics Committee of People’s Hospital of Ganzhou City, and written informed consent was provided by all patients. The human study was performed in accordance with the Declaration of Helsinki.
Cells Culture
Human MDA-MB-231 and MCF-7 BC cells and their PTX-resistant sublines (MDA-MB-231/PTX and MCF-7/PTX) were provided by Procell (Wuhan, China). All cells were routinely propagated in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific, Wesel, Germany) with 10% fetal calf serum (Biowest, Nuaillé, France) and 1% antibiotic (streptomycin and penicillin, Thermo Fisher Scientific). Human non-tumorigenic MCF-10A cell line (Bnbio, Beijing, China) was used as the normal control and cultivated in DMEM/F12 medium (Thermo Fisher Scientific) with 5% heat-inactivated horse serum (Gibco, Wellington, New Zealand) as reported.26 All cells were routinely maintained at 37°C, 5% CO2.
Lentiviral Transduction and Transient Transfection of Cells
CircHIPK3-shRNA (sh-circHIPK3) and nontarget shRNA (sh-NC) lentiviruses were provided by Geneseed (Guangzhou, China) and used to infect MCF-7/PTX cells as per the manufacturing protocols. Cells with positive transduction were selected using 2 µg/mL puromycin (Gibco) over 96 h.
Human circHIPK3 (circ_0000284) sequence incorporated with EcoR I and BamH I sites and the scrambled control sequence were synthesized by BGI (Shenzhen, China) and individually inserted into the pCD5-ciR vector (Geneseed) opened with the two sites to generate circHIPK3 overexpressing plasmid and negative plasmid control (Vector). The encoding region of human HK2 (Accession: NM_001371525.1) was provided by BGI and cloned into BamH I and Xho I sites in the pcDNA3.1 vector (Thermo Fisher Scientific) to create HK2 overexpressing plasmid, and a nontarget control plasmid (pcDNA) was made in the same way. MDA-MB-231/PTX and MCF-7/PTX cells (1 × 105) in 6-well plates were transiently transfected using Lipofectamine 3000 (Thermo Fisher Scientific) with 200 ng of the indicated plasmid, 60 nM of circHIPK3-siRNA (si-circHIPK3#1, si-circHIPK3#2 or si-circHIPK3#3) or nontarget siRNA (si-NC), 30 nM of miR-1286 mimic or negative mimic control (miR-NC mimic), 30 nM of miR-1286 inhibitor (in-miR-1286) or control miRNA inhibitor (in-miR-NC) as per the accompanying guidance. Transfected cells were exposed to 0.5 µM of PTX (except for cell viability assay, Sigma-Aldrich, Taufkirchen, Germany) for 24 h after 24 h or harvested for further analysis after 48 h. All oligonucleotides were provided by Ribobio (Guangzhou, China) and their sequences were in Supplement Table 1.
RNA Preparation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
RNA was prepared from cultured cells (~1 × 1010) and tissue samples using a RNX-Plus Kit (CinnaGen, Tehran, Iran) based on the instructions of manufacturers. cDNA synthesis was conducted with 1 µg of RNA using iSCRIPT cDNA Synthesis Kit (Bio-Rad, Glattbrugg, Switzerland) for circHIPK3 and mRNA quantification or miScript RT Kit (Qiagen, Crawley, UK) for miR-1286 analysis. qRT-PCR with SYBR Green (Qiagen) and designed primers (Supplement Table 1) were performed on the Bio-Rad iQ5 cycler. Results were normalized to β-actin or U6 and were converted to relative expression using the 2−ΔΔCt method.
Ribonuclease R (RNase R) Assay
RNase R assay was done by incubating 2 µg of RNA with 10 U of RNase R for 20 min as per the manufacturing instructions (Epicenter, Stockholm, Sweden).
Subcellular Localization Assay
We isolated cytoplasmic and nuclear RNA from cultured cells (1 × 108) with the Cytoplasmic & Nuclear RNA Purification Kit as recommended by the manufacturers (Norgen Biotek, Thorold, ON, Canada). β-actin and U6 were applied as the cytoplasmic and nuclear controls, respectively.
Measurement of the Half Maximal Inhibitory Concentration (IC50) Value for PTX
Transfected cells were seeded into 96-well plates at 1×104 cells/well 24 h before exposure to PTX at the concentrations of 0.01, 0.1, 0.5, 1, 5, 10 and 20 µM. Twenty-four hours later, cell viability was gauged by the Cell Counting Kit-8 (CCK-8, Genomeditech, Shanghai, China) assay as per the accompanying protocols. The optical density was determined using a plate reader (Thermo Fisher Scientific) at 450 nm. The IC50 value for PTX was evaluated from a plot of the percentage of viable cells versus PTX concentration.
Cell Colony Formation Assay
Approximately 200 treated cells in 2-mL culture medium were plated into each well of 6-well plates. The plates were placed at 37°C in a 5% CO2 incubator. After 2 weeks, the colonies were stained with 0.5% crystal violet (Sigma-Aldrich). The colonies with > 50 cells were scored by counting the triplicate wells for each sample.
Cell Cycle Distribution and Apoptosis Assays
To assess cell cycle distribution, treated cells (1 × 106 per sample) were resuspended in a staining solution containing 40 µg/mL propidium iodide (PI, Thermo Fisher Scientific) and 100 g/mL RNase (TaKaRa, Dalian, China) for 30 min. To measure cell apoptosis, treated cells (1 × 106 per sample) were stained with 5 µL of PI (40 µg/mL) and 10 µL of Annexin V labeled by fluorescein isothiocyanate (Annexin V-FITC, Abcam, Cambridge, UK) for 10 min. The stained cells (1 × 104) were immediately analyzed on the FACSCaliburTM System (BD Biosciences, NY, New York, USA).
Determination of Glucose Consumption and Lactate Production
These assays were performed using the Glucose Uptake Colorimetric Assay Kit and Lactate Colorimetric Assay Kit as per the manufacturing instructions (Biovision, San Francisco, CA, USA), respectively.
Animal Studies
Animal studies were done with 5-week-old female BALB/c nude mice (Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China) following the National Standard of the Care and Use of Laboratory Animals, and our study was approved by the Animal Care and Use Committee and Ethics Committee of People’s Hospital of Ganzhou City. All mice were divided into 4 groups (n = 6 per group): sh-NC + PBS (group I), sh-circHIPK3 + PBS (group II), sh-NC + PTX (group III) and sh-circHIPK3 + PTX (group IV). Approximately 5×106 MCF-7/PTX cells stably transduced with sh-NC or sh-circHIPK3 were injected subcutaneously into the right flanks of nude mice. After 3 days (in the preliminary experiment, our results showed a good effect of inhibiting tumor growth by drug intraperitoneal administration at 3 day after cell injection), the group III and group IV were administered with 3 mg/kg PTX every 3 days by intraperitoneal injection as described,27 and the group I and group II were injected with the same volume PBS (TaKaRa). Tumor volume was periodically monitored and calculated by the formula (length × width2/2). After 21 days cell injection, all mice were euthanized and the tumors were excised for weight and expression analysis. The tumors embedded in paraffin were subjected to immunohistochemistry with rabbit anti-HK2 antibody (ab209847, Abcam; dilution 1:500), as described elsewhere.28
Bioinformatics
The online algorithm CircInteractome was applied to predict the miRNA-binding sites to circHIPK3 at https://circinteractome.nia.nih.gov/. Analysis of the molecular targets of miR-1286 was carried out using the mRNA-predicting software Targetscan at http://www.targetscan.org/vert_71/?tdsourcetag=s_pcqq_aiomsg.
Dual-Luciferase Reporter Assay
The sequence segments of circHIPK3 and HK2 3ʹUTR encompassing the miR-1286 complementary sites or mis-matched target sites were synthesized by BGI and individually cloned into the psiCHECKTM-2 dual-luciferase reporter plasmid (Promega, Charbonnières, France). MDA-MB-231/PTX and MCF-7/PTX cells (1 × 105) were transfected with each reporter construct (200 ng) and either miR-1286 mimic or control mimic at 30 nM. Thirty-six hours post-transfection, luciferase activity was gauged by the dual-luciferase reporter assay (Promega).
RNA Immunoprecipitation (RIP) Assay
Cells (1 × 106) were homogenized in ice RIPA buffer (Thermo Fisher Scientific). Cell lysates were incubated with an antibody against Argonaute 2 (anti-Ago2, #MA5-23515, Invitrogen, Tokyo, Japan; dilution 1:500) or isotype IgG (anti-IgG, #05-4500, Invitrogen; dilution 1:1000) for 3 h at 4°C before adding the Protein A/G Magnetic Agarose Beads (Thermo Fisher Scientific) for 3 h. Beads were harvested, and bound RNA was extracted to measure the levels of circHIPK3, miR-1286 and HK2 mRNA. Bound protein was eluted using RIPA buffer to assess the Ago2 level by Western blot with anti-Ago2 antibody (ab186733, Abcam; dilution 1:1000).
Western Blot for HK2
Protein was prepared from cultured cells (~1 × 108) and tissue samples using ice RIPA buffer containing protease and phosphatase inhibitors (TaKaRa) and quantified by Bradford reagent (Bio-Rad). Protein samples (20 µg) were separated in 4–12% SDS-polyacrylamide gradient gels (Bio-Rad), and resolved proteins were blotted to the Clear Blot membrane-p (ATTO, Tokyo, Japan). The blots were probed overnight at 4°C with an antibody specific for HK2 (#MA5-32952, Invitrogen; dilution 1:1000) or loading control β-actin (ab8226, Abcam; dilution 1:1000). Then, the membranes were incubated with IgG secondary antibody (ab6789, Abcam; dilution 1:10000) for 1 h. Immunopositive bands were visualized with Luminol Reagent (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and the signal was analyzed using the Carestream Molecular Imaging Software (Carestream Health, New Haven, CT, USA).
Statistical Analysis
Unless otherwise noted, the mean ± standard deviation was representative of the average of 3 independent biological replicates. Data were analyzed using the Mann–Whitney U-test, Student’s t-test, or analysis of variance (ANOVA) with Tukey’s post hoc test. The Pearson’s correlation test was applied to analyze the correlation between HK2 mRNA level and circHIPK3 or miR-1286 expression in 25 PTX-resistant BC tissues. The levels of significance were set at *P < 0.05, **P < 0.01 and ***P < 0.001.
Results
CircHIPK3 Was Up-Regulated in PTX-Resistant BC Tissues and Cells
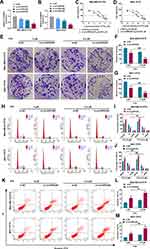
To validate the function of circHIPK3 in PTX-resistant BC, we firstly analyzed its expression pattern in PTX-resistant BC tissues and cells. Analysis of circHIPK3 expression in a panel of human BC tissues showed that circHIPK3 was significantly elevated in BC tissues compared with the adjacent normal breast tissues (Figure 1A). Importantly, circHIPK3 was markedly up-regulated in PTX-resistant BC tissues compared with the corresponding sensitive tissues (Figure 1B). In line with BC tissues, circHIPK3 expression was remarkably increased in MDA-MB-231 and MCF-7 BC cells compared with the non-tumor MCF-10A cells, and PTX-resistant BC cells (MDA-MB-231/PTX and MCF-7/PTX) had higher expression of circHIPK3 compared to their sensitive parents (Figure 1C). To confirm the stability of circHIPK3, we carried out RNase R assays in the two PTX-resistant BC cell lines. Incubation with RNase R led to a striking decrease in the level of HIPK3 linear mRNA, and circHIPK3 was resistant to RNase R (Figure 1D and E). Additionally, subcellular localization assays showed that circHIPK3 was mainly present in the cytoplasm of MDA-MB-231/PTX and MCF-7/PTX cells (Figure 1F and G).
Silencing of circHIPK3 Enhanced Drug Sensitivity of PTX-Resistant BC Cells in vitro
To directly test the functional role of circHIPK3 in drug sensitivity of PTX-resistant BC cells, we designed circHIPK3-siRNA (si-circHIPK3) to inhibit its expression. Transient transfection of si-circHIPK3 dramatically reduced the expression of circHIPK3 in both cell lines compared with the si-NC control (Figure 2A and B). Owing to the most significant inhibition of si-circHIPK3#3 on circHIPK3 expression (Figure 2A and B), we selected it for further investigation. Further analysis showed that PTX-resistant BC cells with circHIPK3 silencing exhibited reduced IC50 value for PTX compared with si-NC control (Figure 2C and D). Furthermore, the reduced expression of circHIPK3 alone remarkably suppressed cell colony formation (Figure 2E–G), cell cycle progression (Figure 2H–J), as well as promoted cell apoptosis (Figure 2K–M). CircHIPK3 silencing alone also impeded glucose consumption (Supplement Figure 1A and B) and lactate production (Supplement Figure 1C and D) in MDA-MB-231/PTX and MCF-7/PTX cells. Intriguingly, simultaneous circHIPK3 silencing and PTX treatment led to a more distinct repression on cell colony formation (Figure 2E–G) and cycle progression (Figure 2H–J), a stronger enhancement on cell apoptosis (Figure 2K–M), as well as a clearer reduction on glucose consumption (Supplement Figure 1A and B) and lactate production (Supplement Figure 1C and D) compared with the si-circHIPK3#3 alone group, indicating that there existed a combinatory effect of PTX and si-circHIPK3#3 on drug sensitivity.
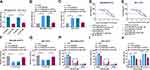
CircHIPK3 Targeted miR-1286 by Directly Binding to miR-1286
To identify novel functional mediators of circHIPK3, we used the computational tool CircInteractome to help identify its targeted miRNAs based on the presence of binding sites. The predicted data showed that circHIPK3 harbored a region that was partially complementary to miR-1286 (Figure 3A). To confirm this, we constructed circHIPK3 wild-type (circHIPK3 WT) or mutant (circHIPK3 MUT) luciferase reporters for standard luciferase assays. Cotransfection of circHIPK3 WT and miR-1286 mimic into the two PTX-resistant cell lines caused lower luciferase activity than cells cotransfected with miR-NC control but circHIPK3 MUT significantly abolished the inhibitory effect of miR-1286 (Figure 3B and C). RIP experiments revealed that the enrichment levels of circHIPK3 and miR-1286 were synchronously augmented by anti-Ago2 antibody (Figure 3D and E). Additionally, miR-1286 was markedly inhibited in BC tissues and cells compared with the normal counterparts, and PTX-resistant cancer tissues and cells had lower expression of miR-1286 compared to the corresponding sensitive controls (Figure 3F–H). To determine whether circHIPK3 could affect miR-1286 expression, we manipulated circHIPK3 expression using si-circHIPK3#3 or circHIPK3 overexpressing plasmid. The transfection efficiency of circHIPK3 overexpressing plasmid was gauged by qRT-PCR (Figure 3I). In contrast, we observed a clear reduction in the levels of the endogenous miR-1286 in circHIPK3-overexpressing cells and an obvious elevation in the expression of miR-1286 in circHIPK3-silencing cells (Figure 3J and K).
miR-1286 Was a Downstream Mediator of circHIPK3 Function in vitro
To elucidate whether miR-1286 was involved in the enhancement of circHIPK3 silencing on drug sensitivity of PTX-resistant BC cells, we knocked down miR-1286 in circHIPK3-silencing cells using miR-1286 inhibitor (in-miR-1286) (Figure 4A–C). Further analysis showed that the down-regulation of miR-1286 highly reversed circHIPK3 silencing-driven IC50 value reduction in both PTX-resistant cell lines (Figure 4D and E). Furthermore, the reduced expression of miR-1286 abolished circHIPK3 silencing-mediated suppression on cell colony formation (Figure 4F and G), cell cycle progression (Figure 4H and I), and promotion on cell apoptosis (Figure 4J) in PTX-resistant BC cells under PTX exposure. Additionally, the reduced level of miR-1286 abrogated the inhibition of glucose consumption (Supplement Figure 2A and B) and lactate production (Supplement Figure 2C and D) of circHIPK3 knockdown in PTX-resistant BC cells exposed to PTX.
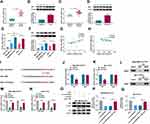
CircHIPK3 Mediated HK2 Expression Through miR-1286
HK2 plays an oncogenic role in breast carcinogenesis and contributes to the development of BC chemoresistance.22,23,25 Our data validated that HK2 mRNA and protein levels were significantly elevated in BC tissues and cells compared with their counterparts, and HK2 mRNA and protein levels were higher in PTX-resistant tissues and cells compared to their sensitive counterparts (Figure 5A–F). Interestingly, we observed that HK2 mRNA level was positively correlated with circHIPK3 expression and inversely correlated with miR-1286 expression in PTX-resistant BC tissues (Figure 5G and H). Using the mRNA-predicting software Targetscan, a putative complementary site for miR-1286 was found within the 3ʹUTR of HK2 (Figure 5I). To establish a direct relationship between miR-1286 and HK2, we cloned the HK2 3ʹUTR segment containing the target region into a luciferase vector. Notably, the luciferase activity of HK2 3ʹUTR reporter was suppressed by miR-1286 mimic (Figure 5J and K). We then generated the site-directed mutation of the seed region and tested it. As expected, the mutant carrying a mutated binding site was refractory to the repression by miR-1286 (Figure 5J and K), demonstrating the validity of the binding site for interaction. Moreover, the enrichment levels of miR-1286 and HK2 mRNA were simultaneously elevated by anti-Ago2 (Figure 5L–N). These data together indicated that miR-1286 directly interacted with the 3ʹUTR of HK2.
We next determined whether circHIPK3 could regulate HK2 expression in the two PTX-resistant BC cell lines. As would be expected, the silencing of circHIPK3 led to a reduction in the level of HK2 protein in both cell lines, and this effect was dramatically abrogated by miR-1286 down-regulation (Figure 5O–Q).
HK2 Was a Functionally Important Target of miR-1286 in Regulating Drug Sensitivity of PTX-Resistant BC Cells in vitro
To elucidate whether miR-1286 affected drug sensitivity of PTX-resistant BC cells by HK2, we overexpressed HK2 using HK2 overexpressing plasmid in cells transfected by miR-1286 mimic. The transfection efficiency of HK2 overexpressing plasmid was verified by Western blot (Figure 6A). In the two PTX-resistant cell lines, the enforced expression of miR-1286 resulted in decreased level of HK2 protein, reinforcing that HK2 was a direct target of miR-1286 (Figure 6B and C). Remarkably, the transfection of HK2 overexpressing plasmid abolished the inhibition of miR-1286 overexpression on HK2 expression in both PTX-resistant BC cell lines (Figure 6B and C). By contrast, the enforced expression of miR-1286 prominently decreased the IC50 value for PTX (Figure 6D and E), suppressed cell colony formation (Figure 6F and G), cell cycle progression (Figure 6H and I), and promoted cell apoptosis (Figure 6J), as well as inhibited glucose consumption (Supplement Figure 3A and B) and lactate production (Supplement Figure 3C and D) in the two PTX-resistant BC cell lines under PTX exposure. However, the restored expression of HK2 remarkably abolished these effects of miR-1286 overexpression (Figure 6D–J, Supplement Figure 3A–D).
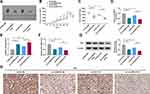
Silencing of circHIPK3 Diminished Tumor Growth and Promoted PTX Sensitivity in vivo
To investigate whether circHIPK3 could influence PTX resistance in vivo, we constructed MCF-7/PTX cells (the in vitro assays revealed that circHIPK3 silencing led to a more significant repression on the proliferation of MCF-7/PTX cells compared with MDA-MB-231/PTX cells under PTX treatment, and we thus selected MCF-7/PTX cells for in vivo assays) stably expressing shRNA-circHIPK3 (sh-circHIPK3) or nontarget shRNA (sh-NC) and injected the cells into the nude mice. In contrast to the corresponding control group, the transduction of sh-circHIPK3 or PTX exposure strikingly repressed tumor growth (Figure 7A–C). Importantly, simultaneous sh-circHIPK3 transduction and PTX exposure led to a more prominent repression on tumor growth (Figure 7A–C). Moreover, sh-circHIPK3 transduction or PTX exposure down-regulated circHIPK3 and HK2 and up-regulated miR-1286 compared with the control group, and simultaneous sh-circHIPK3 transduction and PTX exposure caused a more distinct reduction in the levels of circHIPK3 and HK2, as well as a clearer increase in the expression of miR-1286 in the subcutaneous xenograft tumors (Figure 7D–H).
Discussion
PTX is a first-line chemotherapeutic agent for BC patients, and the development of PTX resistance has become a major therapeutic obstacle in successful BC treatment.4 Overcoming resistance to antineoplastic drugs, including PTX, would represent a significant advance in the treatment of BC. CircRNAs have recently been demonstrated as important modulators in the chemoresistance development of BC.7 Understanding the critical roles of these regulatory molecules would provide a novel basis for molecularly targeted therapeutics. Liu et al found that circ_0006528 depletion sensitized PTX-resistant BC cells to PTX therapy depending on the regulation of miR-1299/CDK8 axis.29 Yang and colleagues showed that circABCB10 silencing enhanced PTX sensitivity of PTX-resistant BC cells by the modulation of the let-7a-5p/DUSP7 axis.27 Considering the important function of circHIPK3 on the chemoresistance development of pancreatic cancer and colorectal cancer,12,13 we here focused on its precise action on PTX resistance of BC. These results reported here uncovered that circHIPK3 silencing promoted PTX sensitivity of PTX-resistant BC cells in vitro and in vivo. Moreover, our findings provided a molecular explanation for circHIPK3-mediated regulation in PTX resistance of BC.
Dysregulation of miR-1286 is reported to be correlated with the overall survival of gastric cancer patients with Helicobacter Pylori infection.30 Hypermethylation of miR-1286 is linked with HPV infection in cervical cells and associated with cervical tumorigenesis.31 Several previous reports had demonstrated the conflicting roles of miR-1286 in human carcinogenesis.19,32–34 These contradictory results might be in part due to the different types of tumors in these reports, where miR-1286 exerted an anti-tumor property in osteosarcoma19,32 and contributed to the progression of cutaneous melanoma33 and lung adenocarcinoma.34 Importantly, miR-1286 expression was remarkably altered by cisplatin or 5-fluorouracil chemotherapy in esophageal cancer cells.35 Our results showed that miR-1286 was down-regulated in PTX-resistant BC, and the enforced expression of miR-1286 could sensitive PTX-resistant cells to PTX treatment. Furthermore, we first uncovered that miR-1286 was a functionally downstream mediator of circHIPK3 in modulating PTX sensitivity of PTX-resistant BC cells.
HK2 has been identified as a tumor driver in numerous cancers, and inhibition of HK2 has been shown as a selective anti-cancer method.36 Work in a number of laboratories has revealed the important involvement of HK2 in the carcinogenesis and chemoresistance of BC.22,23,37 Here, we first identified that HK2 was a functionally important target of miR-1286 in regulating PTX sensitivity of PTX-resistant BC cells. Furthermore, we first established the role of circHIPK3 as a post-transcriptional regulator of HK2 through miR-1286. Enhanced glycolysis, known as the “Warburg effect”, is the main source of energy supply in cancer cells, which contributes to human carcinogenesis.38 By determining glucose consumption and lactate production in PTX-resistant cells, our results suggested that circHIPK3 silencing promoted drug resistance of PTX-resistant BC cells at least partially by regulating cell glycolysis via the miR-1286/HK2 axis. Additionally, the in vivo assays suggested that the enhanced repression of tumor growth under PTX administration might be due to down-regulation of circHIPK3 and HK2 and up-regulation of miR-1286. More studies about the direct regulatory relationship between circHIPK3 and the miR-1286/HK2 axis in drug sensitivity of PTX-resistant BC cells in vivo will be conducted in further work. These findings together with clinical data from patients with PTX-resistant BC showing marked enhancement of circHIPK3 in PTX-resistant tumor tissues provide a rationale for developing circHIPK3 as a therapeutic target for the treatment of PTX-resistant BC.
Similarly, Liu et al underscored that circHIPK3 contributed to gemcitabine resistance by binding to miR-330-5p to induce RASSF1 expression.12 Zhang et al discovered that circHIPK3 exerted a driving effect in the development of oxaliplatin resistance through miR-673.13 Combining with our findings, we envision that the inhibition of circHIPK3 is a starting point for the development of circHIPK3-based therapies against chemoresistance formation.
To conclude, the present study demonstrated that circHIPK3 silencing sensitized PTX-resistant BC cells to PTX therapy through the regulation of the miR-1286/HK2 axis. Our study established the notion that circHIPK3 inhibition might be a good strategy for improving PTX therapy against BC.
Ethics Approval and Consent Participate
Written informed consent was obtained from patients with approval by the Institutional Review Board in People’s Hospital of Ganzhou City.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Ghersi D, Willson ML, Chan MM, et al. Taxane-containing regimens for metastatic breast cancer. Cochrane Database Syst Rev. 2015;2015:Cd003366. doi:10.1002/14651858.CD003366.pub3
3. Megerdichian C, Olimpiadi Y, Hurvitz SA. nab-Paclitaxel in combination with biologically targeted agents for early and metastatic breast cancer. Cancer Treat Rev. 2014;40(5):614–625. doi:10.1016/j.ctrv.2014.02.001
4. Yardley DA. Drug resistance and the role of combination chemotherapy in improving patient outcomes. Int J Breast Cancer. 2013;2013:137414. doi:10.1155/2013/137414
5. Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–692. doi:10.1038/nrg.2016.114
6. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi:10.1038/nrc.2017.99
7. Gao D, Zhang X, Liu B, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9(9):1175–1188. doi:10.2217/epi-2017-0055
8. Dou D, Ren X, Han M, et al. CircUBE2D2 (hsa_circ_0005728) promotes cell proliferation, metastasis and chemoresistance in triple-negative breast cancer by regulating miR-512-3p/CDCA3 axis. Cancer Cell Int. 2020;20(1):454. doi:10.1186/s12935-020-01547-7
9. Zang H, Li Y, Zhang X, et al. Circ-RNF111 contributes to paclitaxel resistance in breast cancer by elevating E2F3 expression via miR-140-5p. Thorac Cancer. 2020;11(7):1891–1903. doi:10.1111/1759-7714.13475
10. Zhang Y, Liu Q, Liao Q. CircHIPK3: a promising cancer-related circular RNA. Am J Transl Res. 2020;12(10):6694–6704.
11. Chen ZG, Zhao HJ, Lin L, et al. Circular RNA CirCHIPK3 promotes cell proliferation and invasion of breast cancer by sponging miR-193a/HMGB1/PI3K/AKT axis. Thorac Cancer. 2020;11(9):2660–2671. doi:10.1111/1759-7714.13603
12. Liu Y, Xia L, Dong L, et al. CircHIPK3 promotes gemcitabine (GEM) resistance in pancreatic cancer cells by sponging miR-330-5p and targets RASSF1. Cancer Manag Res. 2020;12:921–929. doi:10.2147/cmar.s239326
13. Zhang Y, Li C, Liu X, et al. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. 2019;48:277–288. doi:10.1016/j.ebiom.2019.09.051
14. Wang YW, Zhang W, Ma R. Bioinformatic identification of chemoresistance-associated microRNAs in breast cancer based on microarray data. Oncol Rep. 2018;39(3):1003–1010. doi:10.3892/or.2018.6205
15. Wang J, Yang M, Li Y, et al. The role of microRNAs in the chemoresistance of breast cancer. Drug Dev Res. 2015;76(7):368–374. doi:10.1002/ddr.21275
16. Lee JW, Guan W, Han S, et al. MicroRNA-708-3p mediates metastasis and chemoresistance through inhibition of epithelial-to-mesenchymal transition in breast cancer. Cancer Sci. 2018;109(5):1404–1413. doi:10.1111/cas.13588
17. Hong T, Ding J, Li W. miR-7 reverses breast cancer resistance to chemotherapy by targeting MRP1 and BCL2. Onco Targets Ther. 2019;12:11097–11105. doi:10.2147/ott.s213780
18. Santos JC, Lima NDS, Sarian LO, et al. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci Rep. 2018;8(1):829. doi:10.1038/s41598-018-19339-5
19. Yang S, Chen M, Lin CA. Novel lncRNA MYOSLID/miR-1286/RAB13 axis plays a critical role in osteosarcoma progression. Cancer Manag Res. 2019;11:10345–10351. doi:10.2147/cmar.s231376
20. Patra KC, Wang Q, Bhaskar PT, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24(2):213–228. doi:10.1016/j.ccr.2013.06.014
21. Lis P, Dyląg M, Niedźwiecka K, et al. The HK2 dependent “Warburg Effect” and mitochondrial oxidative phosphorylation in cancer: targets for effective therapy with 3-bromopyruvate. Molecules. 2016;21(12):1730. doi:10.3390/molecules21121730
22. Gao Y, Yang Y, Yuan F, et al. TNFα-YAP/p65-HK2 axis mediates breast cancer cell migration. Oncogenesis. 2017;6(9):e383. doi:10.1038/oncsis.2017.83
23. Zhang T, Zhu X, Wu H, et al. Targeting the ROS/PI3K/AKT/HIF-1α/HK2 axis of breast cancer cells: combined administration of Polydatin and 2-Deoxy-d-glucose. J Cell Mol Med. 2019;23(5):3711–3723. doi:10.1111/jcmm.14276
24. Lee HJ, Li CF, Ruan D, et al. Non-proteolytic ubiquitination of Hexokinase 2 by HectH9 controls tumor metabolism and cancer stem cell expansion. Nat Commun. 2019;10(1):2625. doi:10.1038/s41467-019-10374-y
25. He R, Liu H. TRIM59 knockdown blocks cisplatin resistance in A549/DDP cells through regulating PTEN/AKT/HK2. Gene. 2020;747:144553. doi:10.1016/j.gene.2020.144553
26. Wróbel AM, Gregoraszczuk E. Differential effect of methyl-, butyl- and propylparaben and 17β-estradiol on selected cell cycle and apoptosis gene and protein expression in MCF-7 breast cancer cells and MCF-10A non-malignant cells. J Appl Toxicol. 2014;34(9):1041–1050. doi:10.1002/jat.2978
27. Yang W, Gong P, Yang Y, et al. Circ-ABCB10 contributes to paclitaxel resistance in breast cancer through let-7a-5p/DUSP7 axis. Cancer Manag Res. 2020;12:2327–2337. doi:10.2147/cmar.s238513
28. Hatley ME, Patrick DM, Garcia MR, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18(3):282–293. doi:10.1016/j.ccr.2010.08.013
29. Liu G, Zhang Z, Song Q, et al. Circ_0006528 contributes to paclitaxel resistance of breast cancer cells by regulating miR-1299/CDK8 axis. Onco Targets Ther. 2020;13:9497–9511. doi:10.2147/ott.s252886
30. Chu A, Liu J, Yuan Y, et al. Comprehensive analysis of aberrantly expressed ceRNA network in gastric cancer with and without H.pylori infection. J Cancer. 2019;10(4):853–863. doi:10.7150/jca.27803
31. Yao T, Rao Q, Liu L, et al. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in cervical cancer. Virol J. 2013;10(1):175. doi:10.1186/1743-422x-10-175
32. Zhang KQ, Chu XD. GANT61 plays antitumor effects by inducing oxidative stress through the miRNA-1286/RAB31 axis in osteosarcoma. Cell Biol Int. 2020;45(1):61–73. doi:10.1002/cbin.11467
33. Wei X, Gu X, Ma M, et al. Long noncoding RNA HCP5 suppresses skin cutaneous melanoma development by regulating RARRES3 gene expression via sponging miR-12. Onco Targets Ther. 2019;12:6323–6335. doi:10.2147/ott.s195796
34. Gao YW, Ma F, Xie YC, et al. Sp1-induced upregulation of the long noncoding RNA TINCR inhibits cell migration and invasion by regulating miR-107/miR-1286 in lung adenocarcinoma. Am J Transl Res. 2019;11(8):4761–4775.
35. Hummel R, Wang T, Watson DI, et al. Chemotherapy-induced modification of microRNA expression in esophageal cancer. Oncol Rep. 2011;26(4):1011–1017. doi:10.3892/or.2011.1381
36. Garcia SN, Guedes RC, Marques MM. Unlocking the potential of HK2 in cancer metabolism and therapeutics. Curr Med Chem. 2019;26:7285–7322. doi:10.2174/0929867326666181213092652
37. Guo Y, Liang F, Zhao F, et al. Resibufogenin suppresses tumor growth and Warburg effect through regulating miR-143-3p/HK2 axis in breast cancer. Mol Cell Biochem. 2020;466(1–2):103–115. doi:10.1007/s11010-020-03692-z
38. Akram M. Mini-review on glycolysis and cancer. J Cancer Educ. 2013;28(3):454–457. doi:10.1007/s13187-013-0486-9
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.