Back to Journals » OncoTargets and Therapy » Volume 11
SIK1-LNC represses the proliferative, migrative, and invasive abilities of lung cancer cells
Authors Yang L , Xie N, Huang J, Huang H , Xu S, Wang Z, Cai J
Received 11 February 2018
Accepted for publication 30 May 2018
Published 19 July 2018 Volume 2018:11 Pages 4197—4206
DOI https://doi.org/10.2147/OTT.S165278
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Liu Yang,1,* Nianlin Xie,2,* Jingyu Huang,3,* Hu Huang,4,* Shaogan Xu,5 Zhigang Wang,4 Jun Cai6
1Department of Cancer Biotherapy Center, Hubei Cancer Hospital, Wuhan 430079, Hubei, People’s Republic of China; 2Department of Thoracic Surgery, Tangdu Hospital, The Fourth Military Medical University, Xi’an 710038, Shaanxi, People’s Republic of China; 3Department of Thoracic and Cardiovascular Surgery, Zhongnan Hospital of Wuhan University, Wuhan 430071, Hubei, People’s Republic of China; 4Department of Oncology, The 161th Hospital of PLA, Wuhan, Hubei 430010, People’s Republic of China; 5Department of Thoracic Surgery, The 161th Hospital of PLA, Wuhan, Hubei 430010, People’s Republic of China; 6Department of Oncology, First Affiliated Hospital of Yangtze University, Jingzhou 434000, Hubei, People’s Republic of China
*These authors contributed equally to this work
Background: Discussions regarding the correlations between long non-coding RNAs (lncRNAs) and cancers have dominated research in recent years. SIK1-LNC, a type of lncRNA and adjacent to salt-inducible kinases 1 (SIK1), has been found aberrantly expressed in lung cancer. However, its functional role in lung cancer remains largely unknown.
Purpose: In this study, we aimed to explore the association between SIK1-LNC expression and SIK1 in lung cancer cells and further identify the impact of SIK1-LNC on the proliferation, migration invasion of lung cancer cells.
Patients and methods: Of the 30 patients with non-small-cell lung carcinoma from Zhongnan Hospital of Wuhan University, RT-qPCR was performed to detect SIK1 and SIK1-LNC expressions in patients’ samples. Overexpression and knockdown experiments were conducted to analyze the SIK1 and SIK1-LNC expressions in lung cancer cell lines. CCK-8, Brdu, scratch wound-healing, and Transwell assays were respectively employed to evaluate the proliferative, migrative, and invasive abilities of lung cancer cells.
Results: Both SIK1-LNC and SIK1 expression levels were evidently downregulated in 30 lung cancer tissues. SIK1-LNC expression was bound up with clinicopathologic features, including lymph node metastasis and distant metastasis. SIK1 expression showed a positive tendency with SIK1-LNC expression in lung cancer cells. SIK1-LNC exerted a significant repression on cell proliferatiive, miogrative and invasive abilities of lung cancer cells.
Conclusion: Our findings suggested that SIK1-LNC may act as a novel biomarker and therapeutic target for lung cancer.
Keywords: lung cancer, SIK1-LNC, SIK1, proliferation, invasion, metastasis
Introduction
Lung cancer as a major non-communicable disease pertains to one of the most common cancers with the highest incidence and mortality rates in all human malignant tumors.1,2 Generally, the occurrence, development, metastasis, and drug resistance of tumors are closely related to tumor microenvironment.3 Commonly accepted, radiotherapy, chemotherapy, traditional Chinese medicine, and their conjunctions are the approaches for lung cancer treatment.4–7 Despite significant advances made in research into the progression, development, and prognosis of lung cancer over the past few decades, the outcome of most patients afflicted with lung cancer has not remarkably improved. Recently, researchers have disclosed that the vast majorities of human genome are indeed transcribed into various types of non-coding RNAs (ncRNAs) without protein-coding ability, including small ncRNAs (RNA transcript length <200 nt) and long ncRNAs (lncRNAs, RNA transcript length >200 nt).8 Increasing studies have manifested that lncRNAs are widely implicated in a variety of human cancers, including lung cancer,9 colorectal cancer,10 esophageal cancer,11 gastric cancer,12 and osteosarcoma.13 Certainly, numerous lncRNAs have also been found to be related with the occurrence and development of lung cancer. For example, SNHG7 has been reported as an oncogene evidenced by increasing the proliferative, migrative, and invasive abilities, as well as reducing apoptotic potential of lung cancer cells by elevating the expression levels of FAIM2 associated with obesity.14 PCAT615 and CPS1-IT116 were also identified to suppress cell proliferation as an anti-oncogene in human lung cancer. Long intergenetic non-coding RNAs (lincRNAs), belonging to lncRNAs, have been uncovered in the poor-studied genomic regions with many alluring properties, such as associations with various human diseases, tissue-specific expression, and expression changes during development.17–19
As previously reported, lincRNA TCONS_00029157, termed SIK1-LNC in our paper, showed a positive tendency with SIK1 in multiple cancers.20 In our previous work, we found that SIK1-LNC is located at chromosome 21q22.3, ranging from 43,358,055–43,362,688 bp and adjacent to salt-inducible kinases 1 (SIK1) based on the data from the University of California Santa Cruz (UCSC) website.36 Commonly, mounting evidence has revealed that lncRNA can exert its function by targeting either neighboring genes of the same chromosome (in cis) or distantly located genes of the same or different chromosome (in trans).21,22 SIK1, a serine/threonine kinase, belongs to the stress- and energy-sensing AMPK family of kinases, all of which play vital roles in regulation of metabolism and cell growth.23 Recently, emerging evidence has elucidated that SIK1 performs a crucial role in certain human cancers, such as gastric adenocarcinoma24 or epithelial ovarian cancer.25 Particularly in non-small-cell lung cancer, it has been demonstrated that attenuated LKB1-SIK1 signaling enhances epithelial mesenchymal transition and radioresistance of non-small-cell lung cancer cells, which subsequently contributes to the enhanced metastatic potential.9 We, thus, deduced that SIK1 might be a contributor to treatment failure and poor prognosis in non-small-cell lung cancer. In fact, it has been reported that reduced levels of SIK1 are closely correlated with poor outcome in diverse cancers,24 and SIK1 coupled with LKB1 can suppress tumor metastasis.26 However, the expression profile and functional role of SIK1-LNC in lung cancer cells remains largely unclear, which calls for further investigation.
In this study, we first investigated the expression pattern of SIK1-LNC and SIK1 in lung cancer tissues and presented the correlation of SIK1-LNC expression and clinicopathologic features. The association of SIK1-LNC and SIK1 expression in lung cancer cells was also explored. The potentially functional roles of SIK1-LNC in the viability, proliferation, migration, and invasion of lung cancer cells were finally analyzed. The data would provide the theoretical basis for the clinic research of lung cancer in the near future.
Materials and methods
Patient samples
The utilization of samples for this study was approved by the ethical committee of Zhongnan Hospital of Wuhan University. Thirty patients with non-small-cell lung carcinoma who received surgical resection from 2013 to 2014 in Zhongnan Hospital of Wuhan University were selected for this study. All clinical characteristics of patients are listed in Table 1. Of the 30 patients, no one underwent preoperative therapy such as radiotherapy or chemotherapy prior to initial surgery. Also, everyone has signed written informed consent before tumor sample collection. Tissue samples collected at surgery were immediately snap-frozen by liquid nitrogen and stored at −80°C for RNA extraction.
  | Table 1 Correlation between SIK1-LNC expression and clinicopathologic features of lung cancer patients |
Cancer cell lines
Human lung cancer epithelial cell lines (namely BEAS-2B, A549, H1975, and H460) were acquired from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All of them were maintained in RPMI-1640 medium (Gibco BRL, Grand Island, NY, USA) at 37°C in a humidified 5% (v/v) CO2 atmosphere. Ten percent fetal bovine serum, 100 U/mL of penicillin and 100 μg/mL of streptomycin were added into RPMI-1640 medium. Exponentially growing cells were selected for all the experiments in this work.
Quantitative real-time polymerase chain reaction (RT-qPCR) analysis
In accordance with the manufacturer’s protocol, total RNA was extracted from 30 lung cancer tissues and matched adjacent normal tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA was reversely transcribed into complementary DNA using PrimeScript™ One Step RT-PCR kit (Takara Biotechnology Co., Ltd., Dalian, China), and RT-qPCR was performed using the SYBR PrimeScript RT-PCR kit (Takara, Ohstu, Japan) on an Applied Biosystems 7500 Fluorescent Quantitative PCR System (Applied Biosystems, Foster City, CA, USA). The primers were designed and synthesized by GenePharma (Shanghai, China), and all sequences we used are depicted in Table 2. The reaction conditions were designed as follows: an initial single cycle of 30 seconds at 95°C, followed by 40 cycles of 5 seconds at 95°C and finally 34 seconds at 60°C. GAPDH was used as the normal control in this work. The ΔΔCT calculation with CT served as the threshold cycle for quantification of gene expression, and the target gene expression level in the patient was measured as the ratio (target in tumor tissue/target in non-tumorous tissue: R [T/N]).
Western blot analysis
Cells were rinsed with ice-cold PBS harvested and lysated in the lysis buffer (40 nM Tris [pH 7.4], 150 mM NaCl, 10 mM ethylene diamine tetraccetic acid, 10% glycerol, 1% Triton X-100, 10 mM glycerophosphate, 1 mM Na3VO4, and 1 mM phenylmethylsulfonyl fluoride) contained with protease inhibitor (Roche Applied Science, Indianapolis, IN, USA). Proteins were separated on a NuPAGE 4%–12% Bis-Tris Gel (Invitrogen) and then transferred onto a polyvinylidene difluoride membrane (Invitrogen). The membrane was blocked for 60 minutes with 5% skim milk at room temperature, incubated with the primary antibody overnight at 4°C followed by incubation with corresponding secondary antibody. Finally, the membrane was developed by enhancing chemiluminescence solutions (Thermo Fisher Scientific) and detected using the Mini Chemiluminescent Imaging and Analysis System (Sage, China).
Overexpression of SIK1-LNC in lung cancer cells
The SIK1-LNC low-expressed A549 cells were selected to perform the overexpression experiments. The pcDNA SIK1-LNC was constructed by introducing a BamHI-EcoRI fragment into the homologous site in pcDNA3.1, and the introduced BamHI-EcoRI fragment embodied SIK1-LNC cDNA. Following the manufacturer’s instructions, pcDNA SIK1-LNC, and pcDNA control were respectively transfected into A549 cells using Lipofectamine 2000 (Invitrogen). After 48 hours of transfection, cells were applied for RNA isolation, cell proliferation, migration, and invasion assays.
Knockdown of SIK1-LNC and SIK1 in lung cancer cells
Knockdown experiments were carried out in SIK1-LNC high-expressed H460 cells. The small interfering RNAs (siRNAs) for target genes SIK1-LNC (SIK1-LNC si1 and SIK1-LNC si2), negative control siRNA (NC si), and SIK1 (SIK1 si1 and SIK1 si2) were acquired from GenePharma (Shanghai, China), and the sequences are listed in Table 2. Prior to transfection with siRNAs, 5% H460 cells were seeded into 12-well culture plates and maintained for at least 24 hours to achieve a confluence of 30%–50%. According to the manufacturer’s recommendations, siRNA transfections were fulfilled with X-tremeGENE transfection reagent (Roche). Those transfected cells were harvested for RNA isolation, cell proliferation, migration, and invasion assays at 48 hours post-transfection.
Cell viability assays
The viability of transfected cells was assessed by Cell Counting Kit-8 (CCK-8) according to the manufacturer’s recommendation (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Cells transfected with pcDNA or siRNAs were both plated into 96-well plates containing 100 μL of culture medium. Afterwards, CCK-8 (10 μL/well) solution was added to measure cell viability at the designed intervals (0, 24, 48, and 72 hours of incubation). After 2 hours of further incubation at 37°C in 5% CO2, the absorbance of each well was measured at a wavelength of 450 nm by a Model 680 Microplate Reader (Bio-Rad, Richmond, CA, USA).
Cell proliferation assays
The proliferative capacity of the transfected cell was assessed using the bromodeoxyuridine (Brdu) incorporation assay kits (Millipore, Billerica, MA, USA). Briefly, cells (1×104) were seeded in each well of a 96-well plate after 48 hours of transfection. 10 μL of Brdu was subsequently added to each well. After 5 hours incubation, cells were fixed by FixDenat for 30 minutes. Cells were then incubated with anti-Brdu antibody in incubation buffer from Roche for 1 hour at 37°C. Cell nuclei were stained with 1 μM SYTOX Green (S7020, Life Technologies) at room temperature for 15 minutes. In each treatment, cells were washed with PBS at least three times.
Scratch wound-healing assay
The scratch wound healing assay was employed to evaluate the migratory properties of lung cancer cells. Cells at the density of 5×105 cells per well were seeded into plastic 12-well plates and cultured for 12 hours. Uniform wounds were scraped in monolayer cells using a 10 μL pipette tip after being transfected with pcDNA or siRNAs. Wound healing gaps in each group were measured under an optical microscope at 0 and 48 hours.
Cell invasion assays
The invasive properties of transfected cells were examined using transwell chambers invasion assays (8 μm; Corning Incorporated, Corning, NY, USA). After 48 hours of transfection, cells were added into the upper chamber of transwells, and the lower chamber was filled up with complete growth medium. After incubating for 22 hours at 37°C, the non-invading cells were wiped off from the upper surface of membranes, while the invaded cells were stained with 0.5% crystal violet solution. The number of invaded cells pre-well was counted by a microscope ×200 magnification in random fields. All experiments were conducted at least three times.
Statistical analyses
Each presented experiment was performed at least three times, and data were presented as means ± standard deviation (SD). SPSS version 18.0 software (SPSS Inc, Chicago, IL, USA) was used for all statistical analyses. A chi-squared test was employed to evaluate the correlation between SIK1-LNC expression and clinicopathologic features of lung cancer patients. Comparison of different groups was analyzed using the Student’s t-test and a one-way analysis of variance. A value of P<0.05 was considered statistically significant.
Results
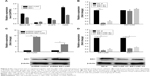
SIK1 and SIK1-LNC were remarkably down-regulated in lung cancer tissue samples
RT-qPCR was performed to detect SIK1 and SIK1-LNC expressions in 30 patient samples. The data suggested that both SIK1 and SIK1-LNC expressions were comparatively down-regulated when compared to those in matched adjacent normal tissues (Figure 1A and B, P<0.05). In addition, the correlation between SIK1-LNC expression and clinicopathologic features of lung cancer patients were also assessed. As shown in Table 1 (P<0.05), SIK1-LNC expression was significantly correlated with lymph node metastasis, distant metastasis. According to correlation analysis (n=30), we found that SIK1-LNC expression was closely associated with SIK1 expression (Figure 1C, P<0.05). By this token, we extrapolated that SIK1 expression may have a positive tendency with SIK1-LNC expression.
SIK1 expression showed a positive tendency with SIK1-LNC expression in lung cell lines
Using RT-qPCR, we examined the SIK1 and SIK1-LNC expressions in four lung cancer cell lines, namely BEAS-2B, A549, H1975, and H460. The results suggested that SIK1-LNC achieved the highest level in H460 and the lowest in A549. So was the case in SIK1 expression (Figure 2A). Low-expressed A549 cells were, therefore, selected to conduct the overexpression experiment, while high-expressed H460 cells were prepared for the knockdown experiment. In SIK1 knockdown experiments, the expression levels of SIK1 were verified, and those of SIK1-LNC remained almost unchanged in cells transfected with either SIK1 si1 or SIK1 si2 when compared to the NC si group (Figure 2B; P<0.05). Next, overexpression and knockdown experiment of SIK1-LNC was conducted and analyzed by RT-qPCR and Western blot assays, respectively. In the SIK1-LNC overexpression experiment, pcDNA SIK1-LNC was constructed and transfected into A549 cells. RT-qPCR analysis suggested that SIK1 and SIK1-LNC expressions at mRNA level in the pcDNA SIK1-LNC group were prominently elevated in contrast with the pcDNA control group. A similar result was obtained in regard to the expression of SIK1 at protein level, as determined by Western blot assay (Figure 2C; P<0.05). Conversely, in the SIK1-LNC knockdown experiment, the expression of SIK1 at both mRNA and protein levels was evidently lower in cells transfected with either SIK1-LNC si1 or SIK1-LNC si2 when compared to the NC si group (Figure 2D; P<0.05). Conjointly, these findings indicated that the SIK1 expression presented a positive tendency with SIK1-LNC expression in lung cancer cells.
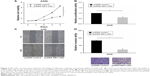
SIK1-LNC overexpression impaired cell viability, proliferative, migrative, and invasive abilities of A549 cells
Cell viability, proliferation, migration, and invasion assays were conducted to investigate the functional role of SIK1-LNC. The pcDNA SIK1-LNC and pcDNA control were constructed for the above experiments. The CCK-8 assay indicated that A549 cell viability in pcDNA SIK1-LNC groups was significantly lower than that in the pcDNA control group (Figure 3A, P<0.05). The Brdu assays showed that the cell proliferation properties were apparently impaired more than 40% in the pcDNA SIK1-LNC group when compared with the pcDNA control group (Figure 3B, P<0.05). Similarly, the wound healing and invasion assays, respectively, indicated that the pcDNA SIK1-LNC group showed significant inhibition on cell migration and invasion in contrast with the pcDNA control group. More concretely, the number of migrated cells in the pcDNA SIK1-LNC group nearly dropped to half as compared to the pcDNA control group and the number of invasive cells in the pcDNA SIK1-LNC group was over three times lower than those in the pcDNA control (Figure 3C and D, P<0.05). Taken together, our findings revealed that SIK1-LNC overexpression could impair cell viability, proliferative, migrative, and invasive abilities of A549 cells.
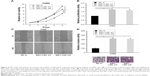
SIK1-LNC knockdown promoted cell viability, proliferative, migrative, and invasive abilities of H460 cells
With the aim of investigation into the functional role of SIK1-LNC in lung cancer cells, knockdown experiments were conducted by transfecting H460 cells with SIK1-LNC si or NC si. It could be observed that the cell viabilities in SIK1-LNC si-transfected groups were gradually higher than those in the NC si group according to CCK-8 assays, and the cell viabilities in SIK1-LNC si-transfected groups had nearly doubled (Figure 4A; P<0.05). The Brdu assays revealed that the cell proliferation abilities of H460 cells were dramatically impaired in SIK1-LNC si-transfected groups when compared with those in the NC si group. Specifically, cell proliferation abilities in SIK1-LNC si-transfected groups were also increased by ~10% (Figure 4B, P<0.05). Likewise, the wound healing and invasion assays manifested a remarkable elevation of cell migration and invasion capacities both in SIK1-LNC si-transfected groups compared with the NC si group. Clearly, cell migration ability increased by ~40% in the SIK1-LNC si-transfected groups, and the cell invasion ability in SIK1-LNC si-transfected groups was over three times more than in the NC si group (Figure 4C and D, P<0.05). Consequently, these results demonstrated that SIK1-LNC knockdown could advance cell viability, proliferative, migrative, and invasive abilities of H460 cells.
Discussion
Recently, mounting studies have demonstrated that lncRNAs are of great significance in cancerogenesis and cancer progression. LncRNAs have become a sharp focus in cancer diagnosis and treatment research.27–29 It has been reported that aberrantly expressed lncRNAs might serve as biomarkers for the early diagnosis of various human cancers, such as lung cancer,9,14–16 hepatocellular carcinoma,30 gastric cancer,12,31,32 mesenchymal glioma,33 and prostate cancer.34,35
In this study, we found a novel tumor suppressor SIK1-LNC is adjacent to protein coding gene SIK1. Commonly, the expression and function of lncRNAs are associated with adjacent protein coding transcripts.21,22 Therein SIK1 has been reported to be positively linked with the treatment of lung cancer,9 and recognized as a potential tumor suppressor in lung cancer. Accordingly, we speculated that SIK1-LNC may be involved in lung cancer development, and its obscure effect on lung cancer spurs us to conduct this research.
In this present study, we found that SIK1-LNC expression was in line with SIK1 expression. Therefore, we deduced that SIK1-LNC may also be implicated in multiple biological processes in lung cancer like cell viability, proliferation, migration, and invasion. To confirm our deduction, the functional experiments of SIK1-LNC in lung cancer cells were performed. We firstly disclosed that SIK1-LNC expression was down-regulated in lung cancer cells and bound up with clinicopathologic features including lymph node metastasis and distant metastasis. This conveyed the possibility of the regulatory role of SIK1-LNC in the occurrence and progression of lung cancer. Subsequently, cell viability, proliferation, migration, and invasion assays were performed to investigate the functional role of SIK1-LNC in lung cancer. The results indicated that the cell viabilities of lung cancer cells were enhanced in the overexpression experiment, but attenuated in the knockdown experiment. Additionally, the results also suggested that overexpression of SIK1-LNC remarkably suppressed the proliferative, migrative, and invasive abilities of A549 cells, while knockdown of SIK1-LNC promoted the cell proliferative, migrative, and invasive abilities of H460 cells.
To our knowledge, this may be the first study concerning the expression pattern and functional role of SIK1-LNC in lung cancer. Our data is likely to provide the first evidence regarding the suppressive role of SIK1-LNC in the viability, proliferation, migration, and invasion of lung cancer cells and its association with prognosis of lung cancer. However, the concrete molecular mechanism of SIK1-LNC in lung cancer has not been investigated in this work. Further studies will include the molecular mechanism of SIK1-LNC in lung cancer and its more evidenced potential for a biomarker and therapeutic target for lung cancer on the basis of this paper.
Conclusions
In summary, this study revealed the suppressive role of SIK1-LNC in lung cancer cells. The data provided a new insight into the occurrence and development of lung cancer. Moreover, SIK1-LNC may be a potential therapeutic target in the treatment of lung cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
Hu CS, Tkebuchava T. SEEDi~(1.0-3.0) strategies for major noncommunicable diseases in China. J Integr Med. 2017;15(4):265. | ||
Nelson A, Sivell S, Noble S, Byrne A, Lester J. Developing an intervention to support lung cancer patients and their clinicians when considering systemic anti-cancer therapy: pact study. Trials. 2013;14(Suppl 1):P8. | ||
Lin WF, Lu JY, Cheng BB, Ling CQ. Progress in research on the effects of traditional Chinese medicine on the tumor microenvironment. J Integr Med. 2017;15(4):282–287. | ||
Tovar I, Expósito J, Jaén J, et al. Pattern of use of radiotherapy for lung cancer: a descriptive study. BMC Cancer. 2014;14(1):697. | ||
Du L, Morgensztern D. Chemotherapy for advanced-stage non-small cell lung cancer. Cancer J. 2015;21(5):366–370. | ||
Zhou ZY, Xu L, Li HG, et al. Chemotherapy in conjunction with traditional Chinese medicine for survival of elderly patients with advanced non-small-cell lung cancer: protocol for a randomized double-blind controlled trial. J Integr Med. 2014;12(3):175–181. | ||
Shen SJ, Zhang YH, Gu XX, Jiang SJ, Xu LJ. Yangfei Kongliu Formula, a compound Chinese herbal medicine, combined with cisplatin, inhibits growth of lung cancer cells through transforming growth factor-β1 signaling pathway. J Integr Med. 2017;15(3):242–251. | ||
Yan H, Bu P. Non-coding RNAs in cancer stem cells. Cancer Lett. 2018;421:121–126. | ||
Yao Y-H, Cui Y, Qiu X-N, et al. Attenuated LKB1-SIK1 signaling promotes epithelial-mesenchymal transition and radioresistance of non–small cell lung cancer cells. Chin J Cancer. 2016;35(1):500–508. | ||
Wen J, Xu J, Sun Q, Xing C, Yin W. Upregulation of long non coding RNA PCAT-1 contributes to cell proliferation, migration and apoptosis in hepatocellular carcinoma. Mol Med Rep. 2016;13(5):4481–4486. | ||
Huang C, Yu Z, Yang H, Lin Y. Increased MALAT1 expression predicts poor prognosis in esophageal cancer patients. Biomed Pharmacother. 2016;83:8–13. | ||
Chen J-F, Wu P, Xia R, et al. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 2018;17(1):6. | ||
Li T, Xiao Y, Huang T. HIF-1α-induced upregulation of lncRNA UCA1 promotes cell growth in osteosarcoma by inactivating the PTEN/AKT signaling pathway. Oncol Rep. 2018;39(3):1072–1080. | ||
She K, Huang J, Zhou H, Huang T, Chen G, He J. lncRNA-SNHG7 promotes the proliferation, migration and invasion and inhibits apoptosis of lung cancer cells by enhancing the FAIM2 expression. Oncol Rep. 2016;36(5):2673–2680. | ||
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Knockdown of long noncoding RNA PCAT6 inhibits proliferation and invasion in lung cancer cells. Oncol Res. 2016;24(3):161–170. | ||
Xiaoguang Z, Meirong L, Jingjing Z, Ruishen Z, Qing Z, Xiaofeng T. Long noncoding RNA CPS1-IT1 suppresses cell proliferation and metastasis in human lung cancer. Oncol Res. 2017;25(3):373–380. | ||
Ching T, Masaki J, Weirather J, Garmire LX. Non-coding yet non-trivial: a review on the computational genomics of lincRNAs. BioData Min. 2015;8:44. | ||
Deniz E, Erman B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct Integr Genomics. 2017;17(2–3):135–143. | ||
Yao Y, Li J, Wang L. Large intervening non-coding RNA HOTAIR is an indicator of poor prognosis and a therapeutic target in human cancers. Int J Mol Sci. 2014;15(10):18985–18999. | ||
Bawa P, Zackaria S, Verma M, et al. Integrative analysis of normal long intergenic non-coding RNAs in prostate cancer. PLoS One. 2015;10(5):e0122143. | ||
Gong WJ, Yin JY, Li XP, et al. Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 2016;37(6):8349–8358. | ||
Preker P, Almvig K, Christensen MS, et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39(16):7179–7193. | ||
Hu Z, Hu J, Shen WJ, Kraemer FB, Azhar S. A novel role of salt-inducible kinase 1 (SIK1) in the post-translational regulation of scavenger receptor class B type 1 activity. Biochemistry. 2015;54(46):6917–6930. | ||
Selvik LK, Rao S, Steigedal TS, et al. Salt-inducible kinase 1 (SIK1) is induced by gastrin and inhibits migration of gastric adenocarcinoma cells. PLoS One. 2014;9(11):e112485. | ||
Wu DD, Chen X, Sun KX, Wang LL, Chen S, Zhao Y. Role of the lncRNA ABHD11-AS1 in the tumorigenesis and progression of epithelial ovarian cancer through targeted regulation of RhoC. Mol Cancer. 2017;16(1):138. | ||
Cheng H, Liu P, Wang ZC, et al. SIK1 couples LKB1 to p53-dependent anoikis and suppresses metastasis. Sci Signal. 2009;2(80):ra35. | ||
Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–R161. | ||
Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. | ||
Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7. | ||
Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6(6):4505–4515. | ||
Dong L, Qi P, Xu MD, et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int J Cancer. 2015;137(5):1128–1135. | ||
Arita T, Ichikawa D, Konishi H, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33(8):3185–3193. | ||
Shi Y, Wang Y, Luan W, et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9(1):e86295. | ||
Ren S, Wang F, Shen J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49(13):2949–2959. | ||
Chakravarty D, Sboner A, Nair SS, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. | ||
SIK1-LNC is located at chromosome 21q22.3, ranging from 43,358,055–43,362,688 bp and adjacent to salt-inducible kinases 1 (SIK1) based on the data from the University of California Santa Cruz (UCSC) Genome Browser database (http://genome.ucsc.edu) [updated July 6, 2018] Available from: http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr21%3A43421112%2D43422005&hgsid=682110025_Fa9GzUJGosJKxxtoM2JQIjRkHxH3. Accessed Dec. 2013. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.