Back to Journals » OncoTargets and Therapy » Volume 13
Significance of Tumor-Infiltrating Immune Cells in the Prognosis of Colon Cancer
Authors Wu D, Ding Y , Wang T, Cui P, Huang L, Min Z, Xu M
Received 18 February 2020
Accepted for publication 5 May 2020
Published 22 May 2020 Volume 2020:13 Pages 4581—4589
DOI https://doi.org/10.2147/OTT.S250416
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Dejun Wu,* Yue Ding,* Tingfeng Wang,* Peng Cui, Liangliang Huang, Zhijun Min, Ming Xu
Department of General Surgery, Shanghai Pudong Hospital, Fudan University Pudong Medical Center, Pudong, Shanghai 201399, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhijun Min; Ming Xu
Department of General Surgery, Shanghai Pudong Hospital, Fudan University Pudong Medical Center, 2800 Gongwei Road, Huinan Town, Pudong, Shanghai 201399, People’s Republic of China
Email [email protected]; [email protected]
Objective: Increasing evidence has indicated an association between immune infiltration in colon cancer and clinical outcomes. The aim of this research is to comprehensively investigate the effect of 22 tumor-infiltrating immune cells (TIICs) on the prognosis of colon cancer patients.
Methods: In our research, CIBERSORT algorithm was used to calculate the proportion of 22 TIICs in 369 colon cancer cases and 39 normal cases from the TCGA cohort. Cox regression analysis was used to analyze the effect of 22 TIICs on the prognosis of colon cancer. Immune risk scoring model was constructed based on the statistical correlation between TIICs subpopulation and survival. Meanwhile, multivariate Cox regression analysis was utilized to investigate whether the immune risk score model was an independent factor for predicting the prognosis of colon cancer. Nomogram was constructed to comprehensively predict the survival rate of colon cancer. P< 0.05 was considered to be statistically significant.
Results: The results of the difference analysis showed that except for 12 TIICs, the remaining immune cells exhibited no differential infiltration between normal and colon cancer tissues (p< 0. 05). Univariate Cox regression analysis revealed 5 immune cells statistically correlated with colon cancer-related survival risk, including B cells naive, B cells memory, monocytes, macrophages M0, macrophages M1 (P< 0.05). In addition, a four-cell based immune risk scoring model was constructed through LASSO Cox regression analysis. KM curve indicated that patients in highrisk were associated with poor outcomes (p< 0.001). ROC curve indicated that the immune risk score model was reliable in predicting survival risk (AUC=0.848). Our model showed satisfying AUC and survival correlation in the validation dataset (3-year over survival (OS) AUC=0.941, 5-year OS AUC=0.865, P=0.022). Furthermore, multivariate Cox regression analysis confirmed that the immune risk score model was an independent factor for predicting the prognosis of colon cancer (hazard ratio (HR) =5.017, 95% confidence interval (CI) =2.336– 10.777; P< 0.001). Ultimately, a nomogram was established to comprehensively predict the survival of colon cancer patients with the results of multivariate Cox regression analysis.
Conclusion: Collectively, tumor-infiltrating immune cells played an essential role in the prognosis of colon cancer. Furthermore, immune risk score was an independent predictive factor of colon cancer, indicating a poor survival.
Keywords: colon cancer, TIICs, prognosis, immune risk score model, nomogram
Introduction
Colon cancer, a principle malignancy of the alimentary canal, ranks third among malignant tumors in terms of morbidity worldwide.1,2 The relevant studies reveal that more than 1 million people developed colon cancer each year, and the disease-specific mortality rate in developed countries is approximately 45%.3 Mortality of colon cancer is on the rise due to changes in diet and lifestyle.4,5 Although colon cancer treatment options (e.g.surgery, chemoradiotherapy, and immunotherapy) have been greatly improved, the 5-year survival rate remains as high as 64.9%.6
It has been extensively documented that the immune system plays an important role in cancer. The immune response is the process of interaction between many specific cells that may affect the clinical outcome of colon cancer.7 It has been reported that the composition and function of tumor-infiltrating immune cells (TIICs) varies with the host immune status and has potential prognostic value. Notably, they could be effectively targeted by drugs and affect the clinical outcomes of patients. CIBERSORT is a gene expression-based deconvolution algorithm that uses a set of barcode gene expression values to characterize immune cell composition. Compared with traditional immunohistochemistry and flow cytometry methods, CIBERSORT algorithm can comprehensively, quickly and accurately infer the relative proportion of 22 types of invasive immune cells in tumors.8 Given the superiority of this method, a large number of studies have recently used this method to investigate the effect of 22 tumor immune cell subtypes on prognosis.9–12
The purpose of this study is to calculate the infiltration extent of 22 immune cells in colon cancer and investigate the effects of 22 TIICs on the prognosis of patients with colon cancer via the CIBERSORT algorithm. Furthermore, an immune risk score model and a nomogram model are constructed to predict the survival of colon cancer.
Materials and Methods
Data Acquisition
We identified and downloaded the transcriptome data of patients with colon cancer from the TCGA database, including 39 cases of normal tissue and 369 cases of tumor tissue. Additionally, relevant clinical information of 369 colon cancer patients were obtained such as age, gender, stage, tumor & Lymph node & metastasis stage, survival status, mutations, microsatellite status, and survival duration. Four GEO datasets (GSE29623, GSE39582, GSE72969, GSE103479) were combined as one independent validation set with 856 cases of colon cancer patients (https://www.ncbi.nlm. nih.gov/geo/). Finally, “limma” package in R software was utilized to correct the transcriptome data we have downloaded.
Assessment of Immune Infiltration
CIBERSORT was a deconvolution algorithm that used 547 tag gene expression values to characterize the composition of immune cells in tissues. In this study, we used this algorithm to estimate the relative proportion of 22 infiltrating immune cells in the corrected transcriptome data of colon cancer. We uploaded the corrected transcriptome data to the CIBERSORT website (http://cibersort.stanford.edu) and set the algorithm to 1000 rows. P < 0.05 was considered to be statistically significant.
Statistical Analysis
All analyses were performed using R 3.6.1. All statistical tests were two-sided, and P value <0.05 was considered statistically significant. Continuous variables that conformed to the normal distribution were compared with the use of independent t test for comparison between groups, while continuous variables with skewed distribution were compared with the Mann–Whitney U-test. The correlation matrix was constructed by R software based on Pearson correlation coefficient. The relationship between immune cell infiltration and overall survival was analyzed through the Kaplan–Meier curve which was evaluated by Log-rank test. Time-dependent ROC curves were used to analyze the sensitivity and specificity of the recurrence prediction model. The univariate regression model was used to analyze the effects of individual variables on survival. The least absolute shrinkage and selection operator (LASSO) Cox regression model was used to confirm independent impact factors associated with survival. The nomogram was constructed with the regression coefficients based on the Cox analysis.
Results
Distribution of Tumor-Infiltrating Immune Cells
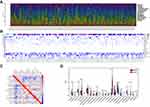
In this study, the dataset included 369 cases of colon cancer samples. The clinicopathological characteristics of these samples are shown in Table 1. The composition of TIIC in colon cancer samples and the correlation between immune cells are shown in Figure 1A and 1B, respectively. We also demonstrated a close relationship between each type of TIIC via Pearson correlation coefficient (Figure 1C). Furthermore, we revealed that there were large differences in the composition of TIIC between normal tissue and colon cancer tissue (Figure 1D). B cells naive, Plasma cells, T cells delta gamma, NK cells activated, Monocytes, Macrophages M2, mast cells resting were decreased whereas T cells CD4 memory activated, T cells follicular helper, NK cells resting, Macrophages M0, mast cells activated were increased in colon cancer tissues compared with healthy bowel tissues. We further investigated the proportion of each TIIL in different pathological stages. We demonstrated that plasma cells, Macrophages M0 and T cells follicular helper were most significantly associated with development of pathological stages (Figure 2).
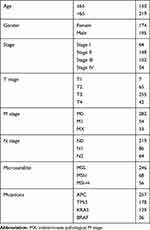 |
Table 1 Clinical Characteristics of Patients Involved in the Study |
LASSO Cox Regression Analysis Was Utilized to Establish an Immune Risk Score
To find a subset of prognosis that was statistically associated with the risk of colon cancer survival, we used a univariate Cox regression method to analyze 22 immune cells. P<0.05 was set as the screening criteria. The results indicated that B cells naive, B cells memory, Monocytes, Macrophages M0, Macrophages M1 were prominently associated with colon cancer survival risk (Table 2). Furthermore, LASSO Cox model was performed to construct an immune risk scoring model based on the foresaid five immune cells (Macrophages M0 was finally eliminated by LASSO Cox model). The distribution of immune risk score, patients’ survival status and 4 immune cells in colon cancer patients was presented in Figure 3C-E, respectively. The correlation between risk scores and clinical indicators is displayed in Figure 3F, and we found the higher immune risk factor was also associated with pathological T and N stage. Each patient received a risk score according to the model. Based on the median value of the risk score, we divided the patients into high-risk and low-risk groups. The ROC curve revealed that the risk model had a good sensitivity and specificity in predicting survival risk (AUC=0.848, Figure 4A). The KM curve suggested that patients in the high-risk group had a poor prognosis (Figure 4B). We also applicated the model in a GEO dataset for validation. As expected, our model showed satisfying AUC in the validation dataset (0.941 for 3-year overall survival (OS) and 0.865 for 5-year OS, Figure 4C). Similarly, patients with the high-risk also showed poor prognosis in validation dataset (Figure 4D).
 |
Table 2 Results of Univariate Cox Regression Analysis for 22 Immune Cells in Colon Cancer Patients |
The Immune Risk Scoring Model Was an Independent Factor for Predicting the Prognosis of Colon Cancer
To explore whether the constructed immune risk scoring model was independent form age, gender, stage, and other clinical pathological parameters, we performed an univariate and multivariate Cox regression analysis for age, gender, stage, TNM and risk score. In univariate Cox model, pathological T, N stage, pathological stage and high immune risk score were associated with poor survival (Figure 5A). In multivariate Cox model, only pathological T stage and immune risk score worked as independent predicted factors (Figure 5B). On the basis of the coefficients of the multivariate Cox regression analysis, we constructed a nomogram to predict patient survival (Figure 5C).
Discussion
As already told, the immune system plays an important role in the development of cancer, which could recognize and kill cancer cells through adaptive immune response defence. Meanwhile, cancer cells could succeed in escaping the recognition of immune cells via a variety of ways. Tumor micro-environment (TME) was composed of cancer cells, stromal cells, chemokines and cytokines, where these cells could communicate and transmit by direct contact or secretion of related cytokines.13 Studies had shown that cancer prognosis was closely related to TME, especially cancer-infiltrating immune cells.14,15 It is evident that different types of cancers had diverse TIICs subpopulation. Even the same pathological type, TIICs subpopulation could be various among patients. Therefore, it was crucial to investigate the immune-infiltrating cell subsets for the evaluation of risk and tumor prognosis.
We conducted a comprehensive and detailed assessment of immune infiltration in colon cancer, based on the deconvolution of data from a large set of samples. The study revealed the specific TIICs that affected the prognosis of patients with colon cancer, and demonstrated the diversity of immune cells in patients with colon cancer. Separate studies have shown that16–19 colon cancer is locally infiltrated with various immune cell subgroups, including Macrophages, dendritic cells, NK cells, initial and memory CD8 + T cells, B cells, and different effector CD4 + T cell subsets, such as Th1 cells, Th2 cells, Th17 cells andregulatory T cells (Treg). These characteristic immune cells constituted an individualized “immune signature map” for patients with early colon cancer, and provided new ideas for the subsequent specific immunotherapy for colon cancer. In our research, we aimed to accurately explore the prognostic value of tumor-infiltrating immune cells in patients with colon cancer. Similarly, the results of the difference analysis showed that except for B cells naive, Plasma cells, T cells delta gamma, T cells CD4 memory activated, T cells follicular helper, NK cells resting, NK cells activated, Monocytes, Macrophages M0, Macrophages M2, mast cells resting, mast cells activated, the remaining immune cells had no differential infiltration between normal and colon cancer tissues (p<0.05).
Additional analysis indicated that B cells naive, B cells memory, Monocytes, Macrophages M0, Macrophages M1 were associated with colon cancer survival risk. Previously, many studies have shown that tumor necrosis factor-α is closely related to the promotion and development of tumors.20,21 Stanilov et al22 have observed that monocytes from patients with advanced cancer secreted significantly more tumor necrosis factor-α than those from patients with early disease. And they believed that monocytes in colon cancer patients are prone to produce higher levels of tumor necrosis factor-α after stimulation, which is related to the survival risk of the disease. Kaler et al recently reported macrophages associated with colon cancer. And they found that macrophages and IL-1 promote Wnt signal transduction in colon cancer cells in a NF-kB-dependent manner. This suggested that the pro-inflammatory cytokines produced by macrophages could enhance the transcription activity of tumor cells and promote the growth of colon cancer cells.23,24 Their results demonstrated that monocytes and macrophages promoted colon cancer cell growth, which was consistent with our findings that increased monocytes infiltration as well as macrophages were associated with poor prognosis of colon cancer.
Furthermore, a LASSO Cox regression method was used to construct an immune risk scoring model for these four immune cells. KM curve indicated that patients in high-risk were associated with poor outcomes (p<0.001). ROC curve indicated that the immune risk scoring model was reliable in predicting survival risk (AUC = 0.848). We conducted a multivariate Cox regression analysis on risk score, age, gender, stage and TNM stages. Results showed that the immune risk score model was an independent factor for predicting the prognosis of colon cancer (hazard ratio (HR) = 5.017, 95%confidence interval (CI) =2.336–10.777; P<0.001). At present, effective biomarkers for predicting the prognosis of colon cancer still lack all over the world.25,26 Clinically, the prognosis was mainly based on the clinical stage and histopathological classification, but in previous studies, it has been found that patients with the same clinical stage and pathological type of colon cancer had different response and prognosis.27,28 Therefore, finding effective prognostic specific TIICs has important clinical value for improving the prognosis of colon cancer. Ultimately, a nomogram was established to comprehensively predict the survival of colon cancer patients with the results of multivariate Cox regression analysis.
Several previous studies have explored the role of immune cells in colon cancer through bioinformatics methods. In 2018, based on the systematic evaluation of the immune status of patients with stage I–III colon cancer, Zhou et al10 compared the immune cell composition of 870 patients with colon cancer and 70 normal controls. A diagnostic model named Diagnostic Immune Risk Score (dIRS) and a prognostic immune risk score (pIRS) were constructed. Unfortunately, they did not evaluate stage 4 colon cancer. It should be noted that we included patients with stage 4 colon cancer and established an immune risk score using four immune cells, which show promising new features for the diagnosis and prognosis prediction of colon cancer. To reveal the important relationship between immune invasion of colon cancer and clinical results, Yongfu et al29 analyzed and evaluated the proportion of tumor-infiltrating immune cell subsets present in 22 colon cancers. However, they did not establish a model to predict survival and prognosis, which was their shortcomings. Ge et al30 explored the expression of immune cells and immune-related genes in the tumor microenvironment of colon cancer. They analyzed the infiltration of 22 immune cells in the TME and the expression of immune-related genes in 404 colon cancer and 40 adjacent non-tumorous tissues. Immune-related genes were selected to establish a prognostic model for TNM staging, but the prediction model was not externally verified in the GEO cohort. We not only used immune-related cells to build an immune risk scoring model, but also applied the model in a GEO dataset for validation. Pleasantly, our model showed satisfying AUC in the validation dataset (0.941 for 3-year OS and 0.865 for 5-year OS). Simultaneously, in Figure 2, we showed the landscape of the distribution of immune cells in different clinical stages and pathological grade colon cancer, which was not described before. Absolutely, our research inevitably had certain limitations. The four validation set data are all from Europe, and thus these results cannot be directly extrapolated to other ethnic groups. The applicability of the model in the Asian population still needs to be verified.
In summary, our study sheds light on the utility of immune cells in the prognosis of colon cancer. The constructed immune risk scoring model is reliable in predicting the prognosis of colon cancer, and this risk scoring model is an independent influencing factor for the prognosis of colon cancer. With the rapid development of high-throughput technology, we have confidence to believe that our immune risk scoring model have great potential in clinical practice. And it may have critical value for exploring new anti-cancer immunodiagnosis and treatment strategies.
Ethical Statement
The authors are accountable for all aspects of the work (if applied, including full data access, integrity of the data and the accuracy of the data analysis) in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Fitzmaurice C, Allen C, Barber RM. A systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3(4):524–548.
3. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386.
4. McGuire S. World Cancer Report 2014. Vol. 2015. Geneva, Switzerland: World Health Organization, international agency for research on cancer, WHO Press; 2016.
5. World Health Organization. Latest Global Cancer Data: Cancer Burden Rises to 18.1 Million New Cases and 9.6 Million Cancer Deaths in 2018. International Agency for Research on Cancer. Geneva: World Health Organization; 2018.
6. Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. The Lancet. 2018;391(10125):1023–1075.
7. Remark R, Becker C, Gomez JE, et al. The non-small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. Am J Respir Crit Care Med. 2015;191(4):377–390.
8. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457.
9. Zhang S, Zhang E, Long J, et al. Immune infiltration in renal cell carcinoma. Cancer Sci. 2019;110(5):1564–1572.
10. Zhou R, Zhang J, Zeng D, et al. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I-III colon cancer. Cancer Immunol Immunother. 2019;68(3):433–442. doi:10.1007/s00262-018-2289-7
11. McDonald KA, Kawaguchi T, Qi Q, et al. Tumor heterogeneity correlates with less immune response and worse survival in breast cancer patients. Ann Surg Oncol. 2019;26(7):2191–2199. doi:10.1245/s10434-019-07338-3
12. Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45(8):860. doi:10.1038/ng.2699
13. Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17(9):559.
14. Bremnes RM, Al-Shibli K, Donnem T, et al. The role of tumor-infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: emphasis on non-small cell lung cancer. J Thor Oncol. 2011;6(4):824–833.
15. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110.
16. Dadabayev R, Sandel MH, Menon AG, et al. Dendritic cells in colorectal cancer correlate with other tumor-infiltrating immune cells. Cancer Immunol Immunother. 2004. 53;11:978–986.
17. Atreya I, Neurath MF. Immune cells in colorectal cancer: prognostic relevance and therapeutic strategies. Expert Rev Anticancer Ther. 2008. 8(4):561–572.
18. Wu Y, Yuan L, Lu Q, et al. Distinctive profiles of tumor-infiltrating immune cells and association with intensity of infiltration in colorectal cancer. Oncol Lett. 2018;15(3):3876–3882.
19. Naito Y, Saito K, Shiiba K, et al. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998;58(16):3491–3494.
20. Hestdal K, Aukrust P, Müller F, et al. Dysregulation of membrane-bound tumor necrosis factor-α and tumor necrosis factor receptors on mononuclear cells in human immunodeficiency virus Type 1 infection: low percentage of p75-tumor necrosis factor receptor positive cells in patients with advance. 1997;90(7):2670–2679.
21. Endo S, Inada K, Wakabayashi G, et al. Tumor Necrosis Factor-a and Tumor Necrosis Factor Receptor I, II Levels in Patients with Severe Burns. Circulation. 2000;26(3):239–244.
22. Stanilov NS, Dobreva ZG, Stanilova SA. Higher TNF-alpha production detected in colorectal cancer patients monocytes. Biotechnol Biotechnol Equip. 2012;26(sup1):107–110.
23. Kaler P, Godasi BN, Augenlicht L, et al. The NF-κB/AKT-dependent induction of wnt signaling in colon cancer cells by macrophages and IL-1β. Cancer Microenviron. 2009;2(1):69–80.
24. Gao L, Zhou Y, Zhou SX, et al. PLD4 promotes M1 macrophages to perform antitumor effects in colon cancer cells. Oncol Rep. 2016.
25. Kehoe J, Khatri VP. Staging and prognosis of colon cancer. Surg Oncol Clin N Am. 15(1):129–146.
26. Zeestraten ECM, Reimers MS, Saadatmand S, et al. Combined analysis of HLA class I, HLA-E and HLA-G predicts prognosis in colon cancer patients. Br J Cancer. 2014;110(2):459–468.
27. West NP, Morris `EJA, Rotimi O, et al. Pathology grading of colon cancer surgical resection and its association with survival: a retrospective observational study. Lancet Oncol. 2008;9(9):857–865.
28. Missiaglia E, Jacobs B, D’ario G, et al. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol. 2014;25(10):1995–2001.
29. Yongfu X, Kang W, He Z, et al. Profiles of immune infiltration in colorectal cancer and their clinical significant: A gene expression-based study. Cancer Med. 2018.
30. Ge P, Wang W, Li L, et al. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of colorectal cancer. Biomed Pharmacother. 2019;109228.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.