Back to Journals » OncoTargets and Therapy » Volume 14
Sevoflurane Suppresses Colon Cancer Cell Malignancy by Regulating circ-PI4KA
Authors Sun S, Wang P, Ren L, Wang H, Zhan Y, Shan S
Received 10 December 2020
Accepted for publication 18 March 2021
Published 20 May 2021 Volume 2021:14 Pages 3319—3333
DOI https://doi.org/10.2147/OTT.S295552
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Suqing Sun, Peng Wang, Lijie Ren, Hongli Wang, Yanli Zhan, Shimin Shan
Department of Anesthesia, Tianjin Fifth Central Hospital, Tianjin, People’s Republic of China
Correspondence: Shimin Shan Department of Anesthesia, Tianjin Fifth Central Hospital, 41 Zhejiang Road, Binhai New Area, Tianjin, 300000, People’s Republic of China
Tel +86 22-6566666
Email [email protected]
Purpose: To explore the effect of SEV on colon cancer cells through circ-PI4KA.
Methods: The RNA level of circular RNA_0062389, microRNA-331-3p and LIM and SH3 protein 1 was determined by quantitative real-time polymerase chain reaction. Protein expression was detected by Western blot. Cell proliferation was investigated by 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide, cell colony formation and 5-ethynyl-29-deoxyuridine assays. Cell apoptosis was demonstrated using Annexin V-fluorescein isothiocyanate/propidium iodide double staining assay. Cell migration and invasion were detected by transwell assay. The target relationship between miR-331-3p and circ-PI4KA or LASP1 was predicted by starBase v2.0 online database, and identified by a dual-luciferase reporter assay. The effects between SEV treatment and circ-PI4KA knockdown on tumor formation were presented by in vivo tumor formation assay.
Results: Circ-PI4KA and LASP1 expressions were dramatically upregulated, while miR-331-3p was downregulated in colon cancer tissues and cells, respectively. SEV exposure significantly decreased the expression of circ-PI4KA and LASP1, but increased miR-331-3p expression. SEV inhibited cell proliferation, migration and invasion, and induced cell apoptosis by regulating circ-PI4KA. Furthermore, circ-PI4KA interacted with miR-331-3p, and miR-331-3p interacted with LASP1. SEV inhibited tumor growth by controlling circ-PI4KA in vivo.
Conclusion: Circ-PI4KA attenuated SEV-treated colon cancer cell malignancy by upregulating LASP1 through binding to miR-331-3p, which provided a new mechanism for studying surgery-mediated therapy of colon cancer.
Keywords: colon cancer, SEV, circ-PI4KA, miR-331-3p, LASP1
Introduction
Colon cancer is the primary trigger of tumor-related mortality worldwide.1 In recent years, the etiopathogenesis of colon cancer has been widely studied, and results suggest that intervening these morbific factors can reduce the incidence of colon cancer.2 Although much progress has been achieved in the diagnosis and therapy of colon cancer, strong abilities of cells in proliferation and metastasis are still a challenge to the treatment of colon cancer. Hence, profoundly elucidating the mechanism of colon cancer process is necessary to improve the 5-year survival rate of patients with colon cancer.
Accumulating data show that anesthetic not only serves as a narcotic but also affects the migratory and invasive abilities of cancer cells.3,4 Sevoflurane (SEV) is a volatile anesthetic which is commonly used in surgical operation. Researchers revealed that SEV suppressed cell proliferation in laryngocarcinoma,5,6 hindered cell proliferation and tumor metastasis in ovarian cancer7 and restrained the proliferation of breast cancer cells.8 However, the molecular mechanism of SEV treatment in colon cancer progression has not been fully demonstrated.
Circular RNA (circRNA) is a noncoding RNA which is more stable than its linear RNA.9 CircRNA is produced by back-splicing with a closed loop.10 Recent evidence has shown that circRNA is enrolled in colon cancer progression.11,12 For instance, circ_0001313 silencing repressed cell viability in colon cancer.13 Zhang et al indicated that circ_0055625 was correlated with tumor metastasis of colon cancer.14 Additionally, Tu et al explained that circ-0001313 absence inhibited cell proliferation and facilitated cell apoptosis in colon cancer.15 However, research on the mechanism of colon cancer regulated by circ_0062389 (circ-PI4KA) is limited. Additionally, previous studies also presented that circRNAs were involved in SEV-mediated cancer progression.16,17 However, the effects of circ-PI4KA on SEV-mediated colon cancer process remain unknown.
MicroRNA (miRNA) is a small RNA with about 22 nucleotides (nts) in size.18 It has been revealed that miRNA exerts function via controlling its associated gene expression through interacting with their 3ʹ-untranslated region (3ʹUTR).19 For example, Sun et al presented that miR-210 silencing facilitated cell viability while suppressed cell cycle arrest via binding to E2F transcription factor 3 in pancreatic cancer.20 Yin et al also unveiled that miR-144 inhibited cell migration and invasion of breast cancer through associating with centrosomal protein 55.21 Another miRNA, miR-331-3p, was reported to act as a tumor promoter or suppressor.22,23 Zhang et al represented that miR-331-3p repressed the migration and invasion of colorectal carcinoma (CRC) cells.24 LIM and SH3 protein 1 (LASP1), an adhesive splice protein, was disclosed to participate in the progression of bowel cancer.25,26 However, in colon cancer, the regulatory mechanism mediated by miR-331-3p and LASP1 is unclear.
Herein, the expression of circ-PI4KA, miR-331-3p and LASP1 was determined in colon cancer tissues and cells. The effects of SEV treatment on the expression of circ-PI4KA, miR-331-3p and LASP1 and the process of colon cancer were revealed. In addition, rescue experiments were performed to identify the mechanism of SEV in colon cancer development.
Materials and Methods
Specimen Acquirement and Preservation
Twenty-seven pairs of human colon cancer tissues and human normal colon tissues were obtained from colon cancer patients from Tianjin Fifth Central Hospital. Tissues were stored in a freezer at −80°C. The Ethics Committee of Tianjin Fifth Central Hospital approved this study. Colon cancer patients signed the written informed consents. Collected colon cancer tissues were divided into 2 groups (low circ-PI4KA expression group and high circ-PI4KA expression group) according to circ-PI4KA expression. The clinicopathologic features of colorectal cancer patients were shown in the Supplementary Table S1. Data displayed that circ-PI4KA expression was not related to gender, age and tumor-node-metastasis (TNM), and correlated with lymph node metastasis and tumor size. Reporting of the study conformed to broad EQUATOR guidelines.
Cell Culture and SEV Treatment
American Type Culture Collection (ATCC, Manassas, VA, USA) supplied human colon cancer cell lines (HCT116, SW620 and SW480), and human normal colonic epithelial cell-line NCM460 was purchased from Otwo Biotech (Shenzhen, China). All the cells were cultivated in Dulbecco’s modified Eagle’s medium (DMEM; Weike Biotechnology, Shanghai, China) supplemented with 10% fetal bovine serum (FBS; Biosun, Shanghai, China) with 1% streptomycin/penicillin (Gibco, Carlsbad, CA, USA) in an incubator with 5% CO2 at 37°C. In addition, NCM 460 cells, SW620 cells or SE480 cells were cultured in DMEM containing 1.7%, 3.4% or 5.1% SEV (Millipore, Bradford, MA, USA) with 0% SEV as a control in vitro based on the description of Sun et al.27
Cell Transfection
Circ-PI4KA overexpression plasmid (circ-PI4KA), miR-331-3p mimic (miR-331-3p), small interfering RNA against circ-PI4KA (si-circ-PI4KA), miR-331-3p inhibitor (anti-miR-331-3p), LASP1 overexpression plasmid (LASP1), small hairpin RNA targeting circ-PI4KA (sh-circ-PI4KA) and controls (vector, miR-NC, si-NC, anti-miR-NC, pcDNA and sh-NC) were synthesized by GENEWIZ (Suzhou, China). Cell transfection was performed using FuGENE6 (Roche, Basel, Switzerland). The sequences were miR-331-3p mimic 5ʹ-GCCCCUGGGCCUAUCCUAGAA-3ʹ, si-circ-PI4KA 5ʹ-TGCAGACCTCCCGACCTCTTT-3ʹ, miR-331-3p inhibitor 5ʹ-UUCUAGGAUAGGCCCAGGGGC-3ʹ, miR-NC 5ʹ-UUUGUACUACACAAAAGUACUG-3ʹ, si-NC 5ʹ-GCTCGACACGTAACCCATA-3ʹ and anti-miR-NC 5ʹ-CAGUACUUUUGUGUAGUACAAA-3ʹ.
3-(4,5)-Dimethylthiahiazo (-z-Y1)-3,5-Di-Phenytetrazoliumromide (MTT) Assay
MTT detection kit (Beyotime, Shanghai, China) was employed to detect cell viability and determine the half maximal inhibitory concentration (IC50) of SEV. In short, NCM cells, SW620 cells or SE480 cells were grown in 96-well plate for 18 h and treated with the desired treatments. After 24 h, cells were incubated with MTT solution (Beyotime) for 4 h in an incubator. Then, dimethyl sulfoxide (Millipore) was incubated with cells at 37°C for 4 h. Cell viability was determined by analyzing the absorbance at 570 nm using a microplate reader (Thermo Labsystems, Waltham, MA, USA). The concentration of SEV causing 50% inhibition of growth (IC50) was determined by assessing the cell viability.
Cell Colony Formation Assay
SW620 and SE480 cells were seeded in 6-well plate (500 cells per well) for 2 weeks after various treatments. Medium was replaced every 3 days. Following that, supernatant was removed and cells were washed with phosphate buffer solution (PBS; Thermo Fisher, Waltham, MA, USA). Crystal violet (Beyotime) was used to stain proliferative colonies for 20 min. Proliferating colonies were photographed, and assessed by counting the number of colonies. Colonies were defined when cell numbers exceeded 50.
5-Ethynyl-29-Deoxyuridine (EDU) Assay
Cell proliferation was further determined by EdU Apollo In Vitro Imaging Kit (Ribobio, Guangzhou, China). Briefly, cells were incubated with EDU solution (Ribobio) for 2 h. Supernatant was discarded, and cells were fixed with methanol (Millipore) for 30 min. After that, cells were incubated with glycine, 0.5% TritonX-100 and Hoechst 33342 reaction buffer, respectively. Finally, samples were assessed by Fluorescence microscopy.
Annexin V-Fluorescein Isothiocyanate (Annexin V-FITC)/Propidium Iodide (PI) Double Staining Assay
Annexin V-FITC apoptosis detection kit (Yeasen Biotech, Shanghai, China) was conducted to detect cell apoptosis. In brief, SW620 and SE480 cells were harvested after digested with trypsin (Thermo Fisher). Then, cells were washed twice using PBS (Thermo Fisher) and precipitated by centrifuging at 260 rpm for 6 min. Following that, binding buffer was carried out to suspend cells, and Annexin V-FITC (Yeasen Biotech) and PI (Yeasen Biotech) were severally used to incubate with cells. Cell apoptosis was unveiled by flow cytometry (BD Biosciences, San Diego, CA, USA).
Transwell Assay
The migration and invasion abilities of SW620 and SE480 cells were determined by transwell chamber without or with Matrigel (Corning, New York, Madison, USA), respectively. Shortly, cells were seeded in the upper chamber of 24-well plate supplemented with serum-free DMEM (Weike Biotechnology). 15% FBS (Biosun) was added to the DMEM and then placed into the lower chamber. After being treated with SEV, circ-PI4KA, vector, anti-miR-331-3p, anti-miR-NC, LASP1 or pcDNA, cells were cultured for 24 h. Chamber was removed from the plate and supernatant was discarded. Methanol (Millipore) was employed to fix cells and crystal violet (Beyotime) was used to dye cells. Results were assessed under a microscope (Olympus, Tokyo, Japan) with 100× magnification.
CircRNA Expression Profile Analysis
In order to find the differently expressed circRNAs in colon cancer tissues without treatment of SEV, the GSE126094 dataset (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE126094) was employed. In brief, the identity document (ID) of screened circRNAs was obtained from circBase online database, and the expression values of samples were determined. Finally, the heatmap was made using GraphPad Prism.
Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
Colon cancer tissues and cells were lysed with TransZol reagent (TransGen Biotech, Beijing, China). And RNAsimple kit (Tiangen, Beijing, China) was performed to extract RNA. Following that, FastKing RT kit (Tiangen) or TQiagen RT kit (Hilden, Germany) was employed to reverse-transcribe RNA into cDNA. In order to assess the expression of circ-PI4KA, miR-331-3p and LASP1, SYBR Green mix (Roche) was carried out. Data were analyzed with the 2−∆∆Ct method. U6 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as references. The sense and antisense primers used in this part were 5’-ACACTCCAGCTGGGGCCCCTGGGCCTATC-3’ and 5’-TGGTGTCGTGGAGTCG-3’ (miR-331-3p) ; 5’-CTCGCTTCGGCAGCACA-3’ and 5’-AACGCTTCACGAATTTGCGT-3’ (U6); 5’-GGTCACCAGGGCTGCTTT-3’ and 5’-GGAAGATGGTGATGGGATT-3’ (GAPDH).
RNase R Treatment Assay
SW620 and SE480 cells were collected, and RNA was isolated using RNAsimple kit (Tiangen). RNA was disposed of with RNase R (Epicentre, Madison, WI, USA) at 37°C for 30 min. Then, RNeasy MinElute Cleaning Kit (Qiagen, Valencia, CA, USA) was used to purify RNA. The stability of circ-PI4KA and linear PI4KA was assessed by detecting their expression via qRT-PCR. Linear PI4KA was selected as a control.
Dual-Luciferase Reporter Assay
The binding sites between miR-331-3p and circ-PI4KA or LASP1 were firstly predicted through starBase v2.0 online database. The wild-type (WT) sequences of circ-PI4KA and LASP1 3ʹUTR containing the binding sites of miR-331-3p were cloned into pmirGLO vector (Promega, Madison, WI, USA), and named as WT-circ-PI4KA and LASP1 3ʹUTR-WT. The sequences bound to miR-331-3p in circ-PI4KA and LASP1 3ʹUTR were mutated and mutant (MUT) circ-PI4KA and LASP1 3ʹUTR were fused into pmirGLO vector (Promega), and defined as MUT-circ-PI4KA and LASP1 3ʹUTR-MUT. Created plasmids were transfected into SW620 and SE480 cells with miR-331-3p mimic or miR-NC using DharmaFECT 4 (Thermo Fisher). Luciferase activities were detected by Dual-Lucy Assay Kit (Solarbio, Beijing, China) with Renilla Luciferase activity as a control.
Western Blot Analysis
Colon cancer tissues and cells were lysed with RIPA buffer (Beyotime). The lysates were mixed with a loading buffer (Thermo Fisher) and then boiled in boiling water. Following that, the lysates were loaded by 12% sodium dodecyl sulfonate-polyacrylamide gel electrophoresis (SDS-PAGE; Beyotime). Then, protein bands were transduced into polyvinylidene fluoride (Millipore), and immersed in 5% nonfat milk (Solarbio) at 4°C for 4 h. And the membranes were incubated with anti-N-cadherin (1:1000; CST, Boston, MA, USA), anti-Vimentin (1:1000; CST), anti-LASP1 (1:1000; CST) and anti-GAPDH (1:1000; CST), respectively, at 4°C for 12 h. Secondary antibody marked with horseradish peroxidase (1:1000; CST) was used to incubate with the membranes at 37°C for 2 h. Bands were visualized with enhanced chemiluminescence (KeyGen, Nanjing, China). GAPDH protein level was used as a control of LASP1 protein level.
In vivo Tumor Formation Assay
BALB/c nude mice were obtained from Charles River (Beijing, China) and fed in pathogen-free condition. Mice were divided into the 3 groups: sh-NC group, sh-NC+SEV group and sh-circ-PI4KA+SEV group (N=8 per group). SW480 cells (4×106) stably transfected with sh-circ-PI4KA or sh-NC were subcutaneously injected into the right back of mice. 7 days later, mice were inhaled 3.4% SEV (Millipore), and tumor volume was measured every 3 days. 22 days later, all mice were killed and tumor weight was detected. For detecting the expression of miR-331-3p and LASP1 in vivo, a part of every tumor was stored at −80°C. The Animal Care and Use Committee of Tianjin Fifth Central Hospital agreed with this study. And the study obeyed the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978).
Data Analysis
All data were obtained based on at least 3 experiments. SPSS 21.0 software (IBM, Somers, NY, USA) was performed to assess the data. Data were showed as means ± standard deviations (SD). Spearman correlation analysis was used to analyze the linear relationship between miR-331-3p and circ-PI4KA or LASP1. A chi-square test was performed to compare the significant differences between low and high circ-PI4KA expression. Pairwise differences were unveiled by two-tailed Student’s t-tests or one-way analysis of variance test. P value < 0.05 was considered statistically significant.
Results
SEV Inhibited Proliferation, Migration and Invasion, and Induced Apoptosis of Colon Cancer Cells
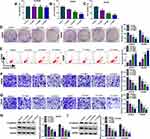
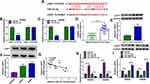
The effects of SEV on colon cancer cell processes were firstly revealed. Before that, the IC50 doses of SEV were detected, and results showed the doses were 4.2% and 3.5% in SW620 and SW480 cells, respectively (Supplementary Figure S1A and B). Then, MTT assay showed that there was no significant (ns) effect on the viability of NCM460 cells after SEV treatment at various concentrations (1.7%, 3.4% and 5.5%) (Figure 1A), whereas SEV treatment suppressed the viability of SW620 and SW480 cells in a dose-dependent manner (Figure 1B and C). Similarly, the colony-forming ability of SW620 and SW480 cells was repressed after exposure to SEV in a dose-dependent manner (Figure 1D). Annexin V-FITC/PI double staining assay demonstrated that SEV treatment promoted cell apoptosis in a dose-dependent fashion in SW620 and SW480 cells (Figure 1E). In addition, it was found that migration and invasion abilities of SW620 and SW480 cells were suppressed by SEV treatment in a concentration-dependent manner (Figure 1F and G). Moreover, the expression of metastasis-related proteins (N-cadherin and Vimentin) was dose-dependently repressed after treatment of SEV (Figure 1H and I). 3.4% SEV was chosen for subsequent experiments as its best inhibitory effect on cell processes. These data demonstrated that SEV could suppress colon cancer cell malignancy.
Circ-PI4KA Expression Was Upregulated in Colon Cancer Tissues and Cells with Association with Poor Survival Rate
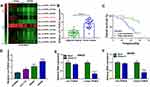
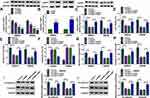
In order to find important circRNAs in colon cancer progression, GSE126094 microarray dataset was employed. Results showed that circRNA_103164 expression was higher in colon cancer tissues than in normal colon tissues when compared with other groups (Figure 2A). Hsa_circ_103164 was named as hsa_circ_0062389 (circ-PI4KA) according to circbank online database (data not shown). QRT-PCR data exhibited that circ-PI4KA was highly expressed in colon cancer tissues (N=27) and HCT-116, SW620 and SW480 cells as compared with paracancerous normal colon tissues and NCM460 cells, respectively (Figure 2B and D). In addition, results unveiled that high circ-PI4KA expression was associated with poor survival rate of colon cancer patients (Figure 2C). Furthermore, RNase R treatment assay presented that circ-PI4KA was more stable than linear PI4KA (Figure 2E and F). The above data suggested that cir-PI4KA might play a vital role in colon cancer progression.
Circ-PI4KA Overexpression Attenuated the Effects of SEV on Colon Cancer Cell Malignancy
Previous study has explained circ_0062389 (circ-PI4KA) contributes to lung cancer progression.28 In order to elucidate whether circ-PI4KA participated in the progression of colon cancer cells mediated by SEV, the effects of circ-PI4KA overexpression on colon cancer progression were firstly determined. Results presented enforced circ-PI4KA expression contributed to cell viability and colony-forming ability of SW620 and SW480 cells (Supplementary Figure S2A and B). EDU assay also showed EDU positive cells were dramatically more in circ-PI4KA group than in the vector group (Supplementary Figure S2C). Transwell assay showed that the migration and invasion of SW620 and SW480 cells were promoted by enforced circ-PI4KA expression (Supplementary Figure S2D and E). These data suggested circ-PI4KA might act as an oncogene in colon cancer development.
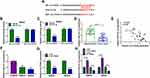
Next, qRT-PCR showed that SEV treatment strikingly downregulated circ-PI4KA expression in a dose-dependent manner (Figure 3A). And circ-PI4KA expression was dramatically upregulated after circ-PI4KA overexpression in SW620 and SW480 cells (Figure 3B), suggesting circ-PI4KA was effective in upregulating circ-PI4KA expression. Subsequently, results presented that circ-PI4KA overexpression abolished the inhibitory effect of SEV on circ-PI4KA expression in SW620 and SW480 cells (Figure 3C). MTT assay demonstrated that circ-PI4KA overexpression hindered the inhibitory effect of SEV treatment on the viability of SW620 and SW480 cells (Figure 3D). Colony formation assay presented that SEV exposure repressed cell colony-forming ability, whereas ectopic circ-PI4KA expression restored this impact in SW620 and SW480 cells (Figure 3E). Furthermore, circ-PI4KA overexpression relieved the promotion of SEV on the apoptosis of SW620 and SW480 cells (Figure 3F). Transwell assay demonstrated that the migration and invasion of SW620 and SW480 cells were inhibited by SEV treatment, whereas these effects were abolished by circ-PI4KA overexpression (Figure 3G and H). Also, the repressive impacts of SEV on the protein expression of N-cadherin and Vimentin were attenuated after overexpression of circ-PI4KA (Figure 3I and J). Our findings suggested that circ-PI4KA was involved in SEV-mediated malignancy repression of colon cancer.
Circ-PI4KA Was Associated with miR-331-3p in Colon Cancer Cells
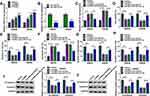
In order to determine the underlying mechanism by which circ-PI4KA affected SEV-mediated malignancy suppression of colon cancer, the miRNA associated with circ-PI4KA was sought. StarBase v2.0 online database showed that circ-PI4KA contained the binding sites (5-’CCCCUGG-3ʹ) of miR-331-3p (Figure 4A). The sequence of circ-PI4KA bound to miR-331-3p was mutated and presented in Figure 4A. Dual-luciferase reporter assay showed that the value of firefly luciferase activity/renilla luciferase activity was dramatically downregulated after WT-circ-PI4KA and miR-331-3p mimic co-transfection, whereas luciferase activity had no obvious change in MUT-circ-PI4KA and miR-331-3p group (Figure 4B and C). Subsequently, qRT-PCR results showed that miR-331-3p expression was notably downregulated in colon cancer tissues and SW620 and SW480 cells compared with control groups (Figure 4D and F). It was found that circ-PI4KA expression was negatively related to miR-331-3p expression (Figure 4E). In addition, the effects of circ-PI4KA mimic or inhibitor on miR-331-3p expression were further revealed. Results firstly showed that circ-PI4KA expression was significantly downregulated after circ-PI4KA knockdown in SW620 and SW480 cells (Figure 4G). It was then disclosed using qRT-PCR that circ-PI4KA knockdown upregulated miR-331-3p expression, and that circ-PI4KA overexpression downregulated miR-331-3p expression (Figure 4H). These results demonstrated that circ-PI4KA was associated with miR-331-3p in SW620 and SW480 cells.
MiR-331-3p Knockdown Abolished the Effects of SEV Treatment on Colon Cancer Process
Given the targeting relationship between circ-PI4KA and miR-331-3p, we continued to explore the effects of miR-331-3p on colon cancer cell processes. As shown in Supplementary Figure S3A and B, miR-331-3p inhibitors contributed to cell proliferation. The migratory and invasive abilities of SW620 and SW480 cells were also enhanced by anti-miR-331-3p (Supplementary Figure S3C and D). These data suggested that miR-331-3p might be implicated in SEV-mediated colon cancer cell malignancy. The effect of miR-331-3p on cell malignancy of colon cancer under SEV exposure was further revealed. QRT-PCR results primarily showed that SEV treatment apparently upregulated miR-331-3p expression in a concentration-dependent fashion (Figure 5A). And miR-331-3p expression was prominently downregulated in SW620 and SW480 cells transfected with anti-miR-331-3p compared with that in SW620 and SW480 cells transfected with anti-miR-NC (Figure 5B). Subsequently, results showed that miR-331-3p inhibitor attenuated SEV-mediated augment on miR-331-3p expression (Figure 5C). Cell viability assay analyzed using MTT showed that miR-331-3p repression restored the inhibitory effect of SEV exposure on cell viability (Figure 5D). Similarly, miR-331-3p depletion restrained the inhibitory effect of SEV incubation on cell colony-forming ability (Figure 5E). In addition, it was revealed that miR-331-3p inhibitor hindered the effect of SEV stimulation on cell apoptosis (Figure 5F). Transwell assay showed that the downregulation of miR-331-3p expression impaired the inhibitory effects of SEV on cell migration and invasion (Figure 5G and H). Also, the inhibitory effects of SEV on the protein expression of N-cadherin and Vimentin were restored by anti-miR-331-3p (Figure 5I and J). Our findings indicated that miR-331-3p was involved in SEV-mediated regulation in colon cancer cell malignancy.
MiR-331-3p Interacted with LASP1 in Colon Cancer Cells
The underlying mechanism of miR-331-3p in SEV-mediated colon cancer cell malignancy was continued to be explored. StarBase v2.0 online database showed that LASP1 3ʹUTR contained the binding sites of miR-331-3p (Figure 6A). And mutant LASP1 3ʹUTR is shown in Figure 6A. In order to identify whether miR-331-3p bound to LASP1 3ʹUTR through the complementary sites between them, dual-luciferase reporter assay was performed. Results showed that luciferase activity was strikingly suppressed in LASP1 3ʹUTR-WT+miR-331-3p mimic group, whereas there was no apparent change in LASP1 3ʹUTR-MUT+miR-331-3p mimic group (Figure 6B and C). Subsequently, it was revealed that LASP1 mRNA and protein expression were dramatically upregulated in colon cancer tissues or SW620 and SW480 cells compared with control groups (Figure 6D–F). LASP1 expression was also unveiled to be negatively correlated with miR-331-3p expression (Figure 6G). Furthermore, the effects of miR-331-3p mimic or inhibitor on LASP1 expression were detected. Results firstly showed that miR-331-3p expression was dramatically upregulated by miR-331-3p mimic and downregulated by miR-331-3p inhibitor (Figure 6H), suggesting miR-331-3p mimic or inhibitor was effective. And Western blot analysis revealed that miR-331-3p mimic apparently downregulated LASP1 protein expression, and that miR-331-3p inhibitor effectively upregulated LASP1 protein expression (Figure 6I). Thus, this evidence proved that miR-331-3p was associated with LASP1 in colon cancer cells.
LASP1 Overexpression Restrained the Effects of SEV Treatment on Colon Cancer Cell Malignancy
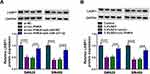
This study continued to explore whether LASP1 participated in the regulation of colon cancer cell malignancy mediated by SEV. The effects of SEV treatment (1.7%, 3.4% and 5.1%) on LASP1 expression were incipiently determined, and results showed that SEV exposure dramatically decreased LASP1 protein expression (Figure 7A). Subsequently, we proved that the overexpression plasmid of LASP1 was successfully built because of LASP1 protein expression was significantly upregulated after LASP1 overexpression (Figure 7B). And results from Western blot analysis exhibited that LASP1 overexpression hindered the inhibitory effect of SEV treatment on LASP1 protein expression (Figure 7C). It was also presented that LASP1 overexpression attenuated the repressive impacts of SEV exposure on the viability and colony-forming ability of SW620 and SW480 cells (Figure 7D and E). Additionally, Annexin V-FITC/PI double staining assay demonstrated that the promotion effect of SEV on cell apoptosis was also reversed after LASP1 overexpression (Figure 7F). Ectopic LASP1 expression blocked the inhibitory effects of SEV exposure on cell migration and invasion in SW620 and SW480 cells (Figure 7G and H). Furthermore, the data from Western blot showed that ectopic LASP1 expression relieved SEV-mediated impacts on the expression of N-cadherin and Vimentin (Figure 7I and J). Thus, these results presented that LASP1 participated in SEV-mediated colon cancer cell malignancy.
Circ-PI4KA Regulated LASP1 Expression by Binding to miR-331-3p
Given the targeting relationship between miR-331-3p and circ-PI4KA or LASP1 as well as the effects of circ-PI4KA, miR-331-3p or LASP1 on colon cancer cell malignancy under SEV exposure, whether circ-PI4KA modulated LASP1 expression by binding to miR-331-3p under SEV exposure was further explored. Data showed that circ-PI4KA knockdown dramatically downregulated LASP1 protein expression, whereas this impact was attenuated by miR-331-3p inhibitor (Figure 8A). Subsequently, the impact of circ-PI4KA on LASP1 expression under SEV treatment was further investigated. Western blot results showed that circ-PI4KA overexpression significantly upregulated LASP1 protein expression under SEV treatment in SW620 and SW480 cells (Figure 8B). These findings suggested that circ-PI4KA regulated LASP1 expression by interacting with miR-331-3p in SEV-induced colon cancer cells.
Circ-PI4KA Knockdown Enhanced the Inhibitory Effects of SEV Treatment on Tumor Formation in vivo
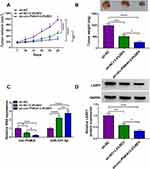
The impacts between circ-PI4KA silencing and SEV treatment on tumor growth in vivo were revealed in this part. Results showed that SEV treatment dramatically repressed tumor volume and weight in vivo, and these effects were enhanced by circ-PI4KA knockdown (Figure 9A and B). Subsequently, the impacts of circ-PI4KA depletion on the expressions of miR-331-3p and LASP1 in vivo were unclosed. QRT-PCR analysis firstly showed that circ-PI4KA knockdown facilitated the inhibitory effect of SEV on circ-PI4KA expression (Figure 9C). And results also displayed that circ-PI4KA silencing enhanced the promotion effect of SEV incubation on miR-331-3p expression (Figure 9C) and the inhibitory effect of that on LASP1 protein expression (Figure 9D). To further determine whether SEV repressed tumor growth by regulating circ-PI4KA, the effects of circ-PI4KA silencing and overexpression on tumor formation were revealed. Results presented that circ-PI4KA knockdown repressed tumor volume and weight (Supplementary Figure S4A and B). Additionally, qRT-PCR data presented that circ-PI4KA expression was apparently downregulated after circ-PI4KA silencing (Supplementary Figure S4C), suggesting sh-circ-PI4KA was effective in knocking down circ-PI4KA expression in vivo. The opposite effect was observed after circ-PI4KA reintroduction (Supplementary Figure S5A-C). Overall, these data showed that circ-PI4KA knockdown enhanced the sensitivity of tumor to SEV.
Discussion
Colon cancer is a common tumor and leads the world in both morbidity and mortality, especially in the elderly.29 SEV, an inhalational hypnotics, has been unveiled to inhibit the proliferation of breast cancer cells8 and the migration of cervical cancer cells.30 In this study, we found that SEV treatment repressed colon cancer cell malignancy. Furthermore, circ-PI4KA overexpression attenuated the impacts of SEV on colon cancer cell malignancy by miR-331-3p/LASP1 axis.
SEV has been showed to regulate bowel cancer process. Yang et al explained that SEV exposure facilitated cell apoptosis and suppressed cell proliferative and invasive abilities in colon cancer.31 Sun et al exhibited that the migratory and invasive abilities of CRC were suppressed by SEV.27 In the current research, in order to study the effects of SEV treatment on colon cancer development, SW620 and SW480 cells were pretreated with SEV (1.7%, 3.4% and 5.1%). Results showed that SEV exposure repressed cell proliferation, migration and invasion, and contributed to cell apoptosis in colon cancer.
CircRNA plays a vital role in colon cancer progression. For instance, circ_000984 absence hindered cell migration and invasion in colon cancer.32 Wang et al indicated that circ_001988 was lowly expressed and its level was related to cell differentiation and invasion in CRC.33 In this study, circ-PI4KA was found to be involved in colon cancer development for the first time. Our findings firstly showed that circ-PI4KA expression was dramatically upregulated in colon cancer tissues and cells. Subsequently, ectopic circ-PI4KA expression contributed to cell proliferation, migration and invasion. Circ-PI4KA overexpression also restored the inhibitory effects of SEV exposure on cell proliferation, migration and invasion, and the proliferation effect of that on cell apoptosis. Furthermore, in vivo assay explained that circ-PI4KA knockdown promoted the inhibitory effects of SEV on tumor formation. All evidences suggested that circ-PI4KA could regulate SEV-mediated colon cancer cell malignancy.
Numerous studies showed that miRNA was involved in the progression of colon cancer via binding to mRNA.34,35 As reported, miR-129-5p was explained to suppress colon cancer progression via interacting with high motility group box protein 1 (HMGB1).36 MiR-195 was found to inhibit cell proliferation and migration through association with wnt family member 3A (WNT3A) in colon cancer.37 In this study, starBase v2.0 online database and dual-luciferase reporter assay were severally performed to predict and identify the binding sites of miR-331-3p in LASP1. Data showed that miR-331-3p bound to LASP1. Zhao et al have demonstrated that miR-331-3p suppresses cell proliferation and facilitates cell apoptosis in CRC,38 suggesting miR-331-3p was an anti-oncogene in CRC process. Wang et al found that miR-145 inhibited cell migration and invasion via interacting with LASP1 in colon cancer,39 which implied that LASP1 promoted colon cancer progression. In the current study, rescue experiments also exhibited that both miR-331-3p inhibitor and LASP1 overexpression abolished the inhibitory impacts of SEV treatment on the proliferative, migratory and invasive abilities of colon cancer cells, and promotion influence of that on cell apoptosis. These findings also demonstrated that miR-331-3p exerted as a suppressor and LASP1 acted as a promoter in colon cancer cell malignancy.
Conclusion
The expressions of circ-PI4KA and LASP1 were dramatically upregulated, and miR-331-3p expression was downregulated in colon cancer tissues and cells. SEV treatment obviously inhibited the expression of circ-PI4KA and LASP1, and increased miR-331-3p expression. Circ-PI4KA restrained the repressive effects of SEV treatment on cell proliferation, migration and invasion, and the stimulatory impact of SEV on cell apoptosis by regulating LASP1 through binding to miR-331-3p in vitro. Furthermore, circ-PI4KA silencing accelerated the inhibitory effects of SEV exposure on tumor growth in vivo. This evidence not only provides a theoretical foundation for the concentration selection of SEV during the surgery of colon cancer cases but also highlights a new direction for researching the application of SEV in the surgery of colon cancer patients.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding authors on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of Tianjin Fifth Central Hospital. Written informed consent was obtained from all enrolled patients.
Consent for Publication
Patients agree to participate in this work.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval for the version to be published; and agreed to be accountable for all aspects of the work.
Funding
No funding was received.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–117. doi:10.3322/caac.21220
2. Gravitz L. Prevention: tending the gut. Nature. 2015;521(7551):S6–S8. doi:10.1038/521S6a
3. Snyder GL, Greenberg S. Effect of anaesthetic technique and other perioperative factors on cancer recurrence. Br J Anaesth. 2010;105(2):106–115. doi:10.1093/bja/aeq164
4. Chen X, Wu Q, You L, Chen S, Zhu M, Miao C. Propofol attenuates pancreatic cancer malignant potential via inhibition of NMDA receptor. Eur J Pharmacol. 2017;795:150–159. doi:10.1016/j.ejphar.2016.12.017
5. Kvolik S, Glavas-Obrovac L, Bares V, Karner I. Effects of inhalation anesthetics halothane, sevoflurane, and isoflurane on human cell lines. Life Sci. 2005;77(19):2369–2383. doi:10.1016/j.lfs.2004.12.052
6. Kvolik S, Dobrosevic B, Marczi S, Prlic L, Glavas-Obrovac L. Different apoptosis ratios and gene expressions in two human cell lines after sevoflurane anaesthesia. Acta Anaesthesiol Scand. 2009;53(9):1192–1199. doi:10.1111/j.1399-6576.2009.02036.x
7. Zhang C, Wang B, Wang X, Sheng X, Cui Y. Sevoflurane inhibits the progression of ovarian cancer through down-regulating stanniocalcin 1 (STC1). Cancer Cell Int. 2019;19(1):339. doi:10.1186/s12935-019-1062-0
8. Liu J, Yang L, Guo X, et al. Sevoflurane suppresses proliferation by upregulating microRNA-203 in breast cancer cells. Mol Med Rep. 2018;18(1):455–460. doi:10.3892/mmr.2018.8949
9. Shen B, Wang Z, Li Z, Song H, Ding X. Circular RNAs: an emerging landscape in tumor metastasis. Am J Cancer Res. 2019;9(4):630–643.
10. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
11. Zhang Q, Zhang C, Ma JX, Ren H, Sun Y, Xu JZ. Circular RNA PIP5K1A promotes colon cancer development through inhibiting miR-1273a. World J Gastroenterol. 2019;25(35):5300–5309. doi:10.3748/wjg.v25.i35.5300
12. Zheng X, Chen L, Zhou Y, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18(1):47. doi:10.1186/s12943-019-1010-6
13. Wang L, Peng X, Lu X, Wei Q, Chen M, Liu L. Inhibition of hsa_circ_0001313 (circCCDC66) induction enhances the radio-sensitivity of colon cancer cells via tumor suppressor miR-338-3p: effects of cicr_0001313 on colon cancer radio-sensitivity. Pathol Res Pract. 2019;215(4):689–696. doi:10.1016/j.prp.2018.12.032
14. Zhang J, Liu H, Zhao P, Zhou H, Mao T. Has_circ_0055625 from circRNA profile increases colon cancer cell growth by sponging miR-106b-5p. J Cell Biochem. 2019;120(3):3027–3037. doi:10.1002/jcb.27355
15. Tu FL, Guo XQ, Wu HX, et al. Circ-0001313/miRNA-510-5p/AKT2 axis promotes the development and progression of colon cancer. Am J Transl Res. 2020;12(1):281–291.
16. He J, Zhao H, Liu X, et al. Sevoflurane suppresses cell viability and invasion and promotes cell apoptosis in colon cancer by modulating exosome-mediated circ-HMGCS1 via the miR-34a-5p/SGPP1 axis. Oncol Rep. 2020;44(6):2429–2442. doi:10.3892/or.2020.7783
17. Xu W, Xue R, Xia R, et al. Sevoflurane impedes the progression of glioma through modulating the circular RNA has_circ_0012129/miR-761/TGIF2 axis. Eur Rev Med Pharmacol Sci. 2020;24(10):5534–5548. doi:10.26355/eurrev_202005_21339
18. Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136(4):586–591. doi:10.1016/j.cell.2009.02.005
19. Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. doi:10.1016/j.cell.2007.12.024
20. Sun FB, Lin Y, Li SJ, Gao J, Han B, Zhang CS. MiR-210 knockdown promotes the development of pancreatic cancer via upregulating E2F3 expression. Eur Rev Med Pharmacol Sci. 2018;22(24):8640–8648. doi:10.26355/eurrev_201812_16628
21. Yin Y, Cai J, Meng F, Sui C, Jiang Y. MiR-144 suppresses proliferation, invasion, and migration of breast cancer cells through inhibiting CEP55. Cancer Biol Ther. 2018;19(4):306–315. doi:10.1080/15384047.2017.1416934
22. Epis MR, Giles KM, Candy PA, Webster RJ, Leedman PJ. miR-331-3p regulates expression of neuropilin-2 in glioblastoma. J Neurooncol. 2014;116(1):67–75. doi:10.1007/s11060-013-1271-7
23. Jin W, Zhong N, Wang L, Yu J, Yin F, Zhang K. MiR-331-3p Inhibition of the Hepatocellular Carcinoma (HCC) Bel-7402 Cell Line by Down-Regulation of E2F1. J Nanosci Nanotechnol. 2019;19(9):5476–5482. doi:10.1166/jnn.2019.16535
24. Zhang H, Wang R, Wang M. miR-331-3p suppresses cell invasion and migration in colorectal carcinoma by directly targeting NRP2. Oncol Lett. 2019;18(6):6501–6508. doi:10.3892/ol.2019.11029
25. Niu Y, Shao Z, Wang H, et al. LASP1-S100A11 axis promotes colorectal cancer aggressiveness by modulating TGFβ/Smad signaling. Sci Rep. 2016;6:26112. doi:10.1038/srep26112
26. Zhou R, Shao Z, Liu J, et al. COPS5 and LASP1 synergistically interact to downregulate 14-3-3σ expression and promote colorectal cancer progression via activating PI3K/AKT pathway. Int J Cancer. 2018;142(9):1853–1864. doi:10.1002/ijc.31206
27. Sun SQ, Ren LJ, Liu J, Wang P, Shan SM. Sevoflurane inhibits migration and invasion of colorectal cancer cells by regulating microRNA-34a/ADAM10 axis. Neoplasma. 2019;66(6):887–895. doi:10.4149/neo_2018_181213N962
28. She Y, Han Y, Zhou G, Jia F, Yang T, Shen Z. hsa_circ_0062389 promotes the progression of non-small cell lung cancer by sponging miR-103a-3p to mediate CCNE1 expression. Cancer Genet. 2020;241:12–19. doi:10.1016/j.cancergen.2019.12.004
29. Schonberg MA, Breslau ES, Hamel MB, Bellizzi KM, McCarthy EP. Colon cancer screening in U.S. adults aged 65 and older according to life expectancy and age.. J Am Geriatr Soc. 2015;63(4):750–756. doi:10.1111/jgs.13335
30. Ding J, Zhang L, Zeng S, Feng T. Clinically relevant concentration of sevoflurane suppresses cervical cancer growth and migration through targeting multiple oncogenic pathways. Biochem Biophys Res Commun. 2019;514(4):1179–1184. doi:10.1016/j.bbrc.2019.05.082
31. Yang X, Zheng YT, Rong W. Sevoflurane induces apoptosis and inhibits the growth and motility of colon cancer in vitro and in vivo via inactivating Ras/Raf/MEK/ERK signaling. Life Sci. 2019;239:116916. doi:10.1016/j.lfs.2019.116916
32. Xu XW, Zheng BA, Hu ZM, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8(53):91674–91683. doi:10.18632/oncotarget.21748
33. Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–16025.
34. Zeng M, Zhu L, Li L, Kang C. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol Biol Lett. 2017;22(1):12. doi:10.1186/s11658-017-0041-5
35. Teng Y, Ren Y, Hu X, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8(1):14448. doi:10.1038/ncomms14448
36. Wu Q, Meng WY, Jie Y, Zhao H. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol. 2018;233(9):6750–6757. doi:10.1002/jcp.26383
37. Li B, Wang S, Wang S. MiR-195 suppresses colon cancer proliferation and metastasis by targeting WNT3A. Mol Genet Genomics. 2018;293(5):1245–1253. doi:10.1007/s00438-018-1457-y
38. Zhao D, Sui Y, Zheng X. MiR-331-3p inhibits proliferation and promotes apoptosis by targeting HER2 through the PI3K/Akt and ERK1/2 pathways in colorectal cancer. Oncol Rep. 2016;35(2):1075–1082. doi:10.3892/or.2015.4450
39. Wang W, Ji G, Xiao X, et al. Epigenetically regulated miR-145 suppresses colon cancer invasion and metastasis by targeting LASP1. Oncotarget. 2016;7(42):68674–68687. doi:10.18632/oncotarget.11919
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.